Discovery of AG-270, a First-in-Class Oral MAT2A Inhibitor for the Treatment of Tumors with Homozygous MTAP Deletion.
Konteatis, Z., Travins, J., Gross, S., Marjon, K., Barnett, A., Mandley, E., Nicolay, B., Nagaraja, R., Chen, Y., Sun, Y., Liu, Z., Yu, J., Ye, Z., Jiang, F., Wei, W., Fang, C., Gao, Y., Kalev, P., Hyer, M.L., DeLaBarre, B., Jin, L., Padyana, A.K., Dang, L., Murtie, J., Biller, S.A., Sui, Z., Marks, K.M.(2021) J Med Chem 64: 4430-4449
- PubMed: 33829783
- DOI: https://doi.org/10.1021/acs.jmedchem.0c01895
- Primary Citation of Related Structures:
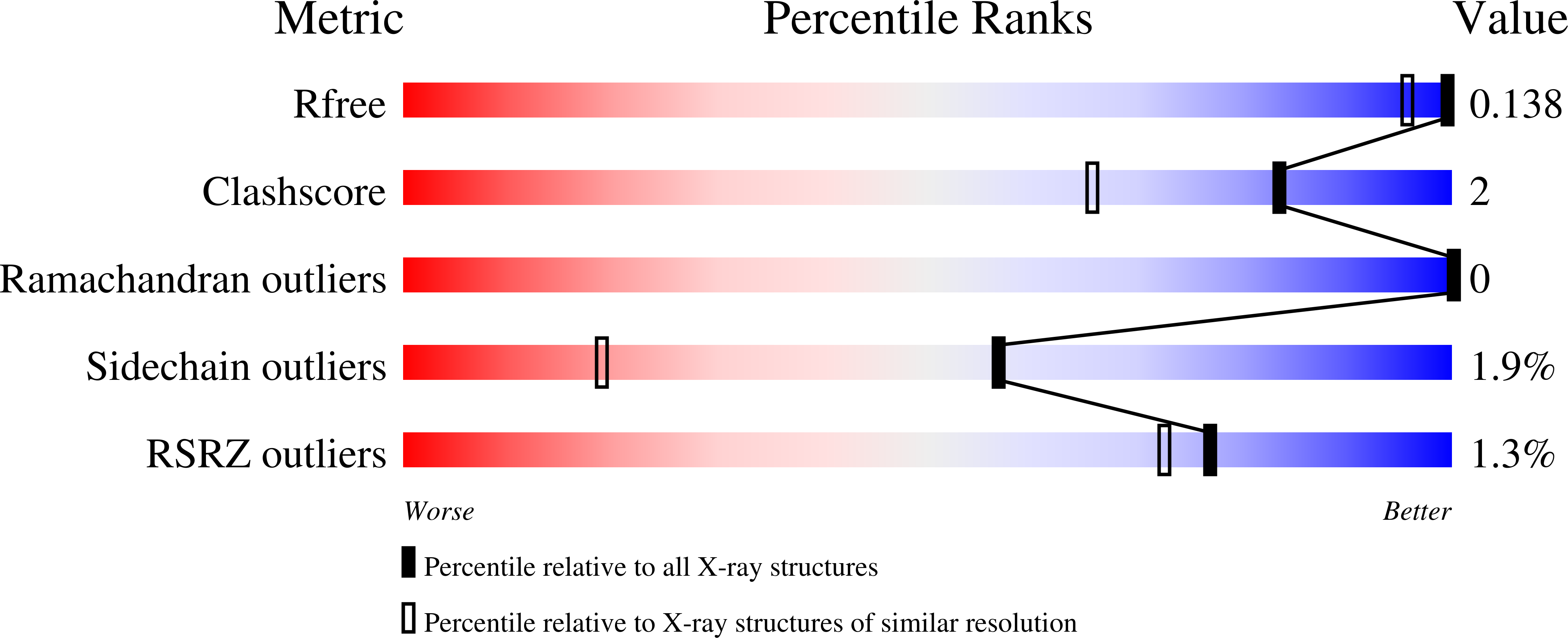
7KCC, 7KCE, 7KCF, 7KDA, 7KDB - PubMed Abstract:
The metabolic enzyme methionine adenosyltransferase 2A (MAT2A) was recently implicated as a synthetic lethal target in cancers with deletion of the methylthioadenosine phosphorylase ( MTAP ) gene, which is adjacent to the CDKN2A tumor suppressor and codeleted with CDKN2A in approximately 15% of all cancers. Previous attempts to target MAT2A with small-molecule inhibitors identified cellular adaptations that blunted their efficacy. Here, we report the discovery of highly potent, selective, orally bioavailable MAT2A inhibitors that overcome these challenges. Fragment screening followed by iterative structure-guided design enabled >10 000-fold improvement in potency of a family of allosteric MAT2A inhibitors that are substrate noncompetitive and inhibit release of the product, S -adenosyl methionine (SAM), from the enzyme's active site. We demonstrate that potent MAT2A inhibitors substantially reduce SAM levels in cancer cells and selectively block proliferation of MTAP -null cells both in tissue culture and xenograft tumors. These data supported progressing AG-270 into current clinical studies (ClinicalTrials.gov NCT03435250).
Organizational Affiliation:
Agios Pharmaceuticals, Inc., 88 Sidney Street, Cambridge, Massachusetts 02139, United States.