Structure, Immunogenicity, and Conformation-Dependent Receptor Binding of the Postfusion Human Metapneumovirus F Protein.
Huang, J., Chopra, P., Liu, L., Nagy, T., Murray, J., Tripp, R.A., Boons, G.J., Mousa, J.J.(2021) J Virol 95: e0059321-e0059321
- PubMed: 34160259
- DOI: https://doi.org/10.1128/JVI.00593-21
- Primary Citation of Related Structures:
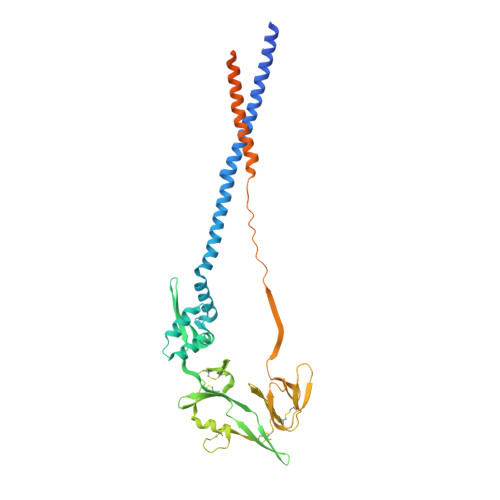
7M0I - PubMed Abstract:
Human metapneumovirus (hMPV) is an important cause of acute viral respiratory infection. As the only target of neutralizing antibodies, the hMPV fusion (F) protein has been a major focus for vaccine development and targeting by drugs and monoclonal antibodies (MAbs). While X-ray structures of trimeric prefusion and postfusion hMPV F proteins from genotype A, and monomeric prefusion hMPV F protein from genotype B have been determined, structural data for the postfusion conformation for genotype B is lacking. We determined the crystal structure of this protein and compared the structural differences of postfusion hMPV F between hMPV A and B genotypes. We also assessed the receptor binding properties of the hMPV F protein to heparin and heparan sulfate (HS). A library of HS oligomers was used to verify the HS binding activity of hMPV F, and several compounds showed binding to predominantly prefusion hMPV F, but had limited binding to postfusion hMPV F. Furthermore, MAbs to antigenic sites III and the 66-87 intratrimeric epitope block heparin binding. In addition, we evaluated the efficacy of postfusion hMPV B2 F protein as a vaccine candidate in BALB/c mice. Mice immunized with hMPV B2 postfusion F protein showed a balanced Th1/Th2 immune response and generated neutralizing antibodies against both subgroup A2 and B2 hMPV strains, which protected the mice from hMPV challenge. Antibody competition analysis revealed the antibodies generated by immunization target two known antigenic sites (III and IV) on the hMPV F protein. Overall, this study provides new characteristics of the hMPV F protein, which may be informative for vaccine and therapy development. IMPORTANCE Human metapneumovirus (hMPV) is an important cause of viral respiratory disease. In this paper, we report the X-ray crystal structure of the hMPV fusion (F) protein in the postfusion conformation from genotype B. We also assessed binding of the hMPV F protein to heparin and heparan sulfate, a previously reported receptor for the hMPV F protein. Furthermore, we determined the immunogenicity and protective efficacy of postfusion hMPV B2 F protein, which is the first study using a homogenous conformation of the protein. Antibodies generated in response to vaccination give a balanced Th1/Th2 response and target two previously discovered neutralizing epitopes.
Organizational Affiliation:
Department of Infectious Diseases, College of Veterinary Medicine, University of Georgiagrid.213876.9, Athens, Georgia, USA.