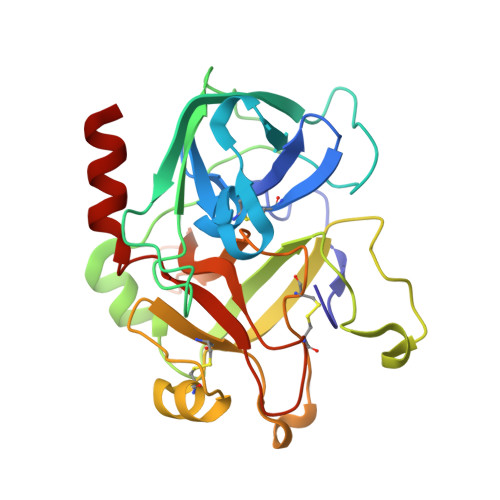
Identification of a substrate-like cleavage-resistant thrombin inhibitor from the saliva of the flea Xenopsylla cheopis.
Lu, S., Tirloni, L., Oliveira, M.B., Bosio, C.F., Nardone, G.A., Zhang, Y., Hinnebusch, B.J., Ribeiro, J.M., Andersen, J.F.(2021) J Biol Chem 297: 101322-101322
- PubMed: 34688666
- DOI: https://doi.org/10.1016/j.jbc.2021.101322
- Primary Citation of Related Structures:
7MJ5 - PubMed Abstract:
The salivary glands of the flea Xenopsylla cheopis, a vector of the plague bacterium, Yersinia pestis, express proteins and peptides thought to target the hemostatic and inflammatory systems of its mammalian hosts. Past transcriptomic analyses of salivary gland tissue revealed the presence of two similar peptides (XC-42 and XC-43) having no extensive similarities to any other deposited sequences. Here we show that these peptides specifically inhibit coagulation of plasma and the amidolytic activity of α-thrombin. XC-43, the smaller of the two peptides, is a fast, tight-binding inhibitor of thrombin with a dissociation constant of less than 10 pM. XC-42 exhibits similar selectivity as well as kinetic and binding properties. The crystal structure of XC-43 in complex with thrombin shows that despite its substrate-like binding mode, XC-43 is not detectably cleaved by thrombin and that it interacts with the thrombin surface from the enzyme catalytic site through the fibrinogen-binding exosite I. The low rate of hydrolysis was verified in solution experiments with XC-43, which show the substrate to be largely intact after 2 h of incubation with thrombin at 37 °C. The low rate of XC-43 cleavage by thrombin may be attributable to specific changes in the catalytic triad observable in the crystal structure of the complex or to extensive interactions in the prime sites that may stabilize the binding of cleavage products. Based on the increased arterial occlusion time, tail bleeding time, and blood coagulation parameters in rat models of thrombosis XC-43 could be valuable as an anticoagulant.
Organizational Affiliation:
Laboratory of Malaria and Vector Research, National Institute of Allergy and Infectious Diseases, Bethesda, Maryland, USA.