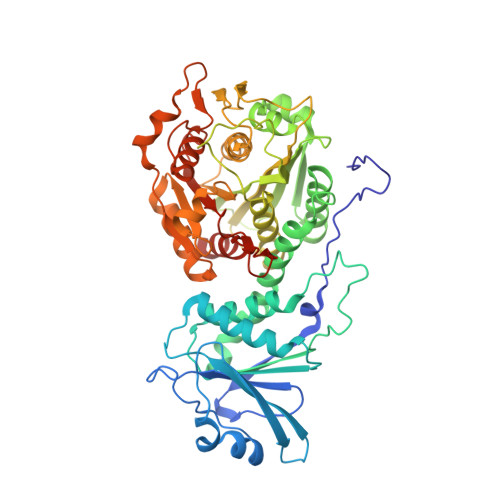
A metal ion-dependent conformational switch modulates activity of the Plasmodium M17 aminopeptidase.
Webb, C.T., Yang, W., Riley, B.T., Hayes, B.K., Sivaraman, K.K., Malcolm, T.R., Harrop, S., Atkinson, S.C., Kass, I., Buckle, A.M., Drinkwater, N., McGowan, S.(2022) J Biol Chem 298: 102119-102119
- PubMed: 35691342
- DOI: https://doi.org/10.1016/j.jbc.2022.102119
- Primary Citation of Related Structures:
7SRV, 7T3V - PubMed Abstract:
The metal-dependent M17 aminopeptidases are conserved throughout all kingdoms of life. This large enzyme family is characterized by a conserved binuclear metal center and a distinctive homohexameric arrangement. Recently, we showed that hexamer formation in Plasmodium M17 aminopeptidases was controlled by the metal ion environment, although the functional necessity for hexamer formation is still unclear. To further understand the mechanistic role of the hexameric assembly, here we undertook an investigation of the structure and dynamics of the M17 aminopeptidase from Plasmodium falciparum, PfA-M17. We describe a novel structure of PfA-M17, which shows that the active sites of each trimer are linked by a dynamic loop, and loop movement is coupled with a drastic rearrangement of the binuclear metal center and substrate-binding pocket, rendering the protein inactive. Molecular dynamics simulations and biochemical analyses of PfA-M17 variants demonstrated that this rearrangement is inherent to PfA-M17, and that the transition between the active and inactive states is metal dependent and part of a dynamic regulatory mechanism. Key to the mechanism is a remodeling of the binuclear metal center, which occurs in response to a signal from the neighboring active site and serves to moderate the rate of proteolysis under different environmental conditions. In conclusion, this work identifies a precise mechanism by which oligomerization contributes to PfA-M17 function. Furthermore, it describes a novel role for metal cofactors in the regulation of enzymes, with implications for the wide range of metalloenzymes that operate via a two-metal ion catalytic center, including DNA processing enzymes and metalloproteases.
Organizational Affiliation:
Biomedicine Discovery Institute, Department of Microbiology, Monash University, Clayton Melbourne, VIC, Australia.