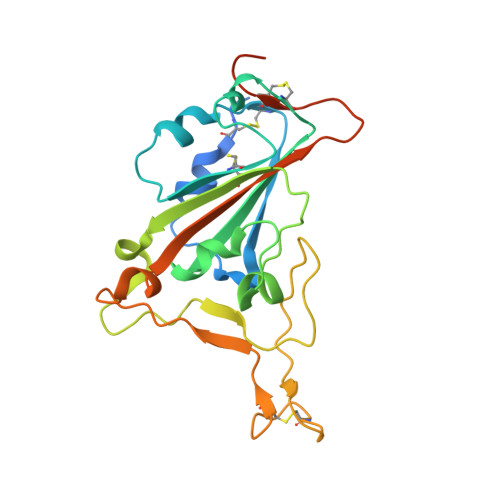
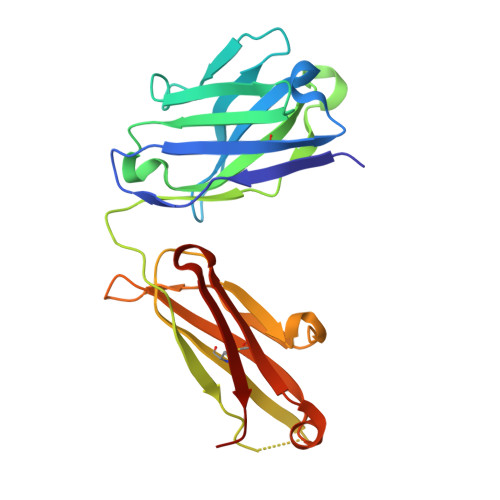
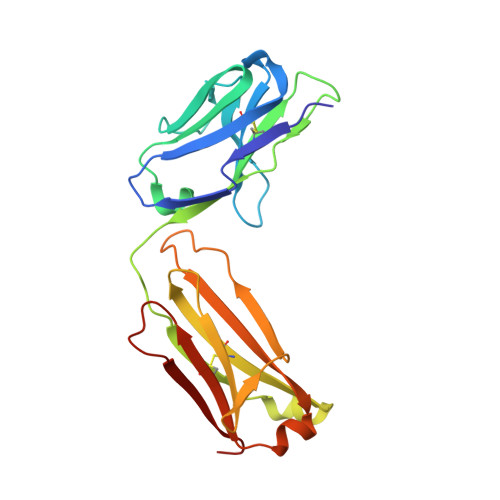
The molecular basis of the neutralization breadth of the RBD-specific antibody CoV11.
Tolbert, W.D., Chen, Y., Sun, L., Benlarbi, M., Ding, S., Manickam, R., Pangaro, E., Nguyen, D.N., Gottumukkala, S., Cote, M., Gonzalez, F.J., Finzi, A., Tehrani, Z.R., Sajadi, M.M., Pazgier, M.(2023) Front Immunol 14: 1178355-1178355
- PubMed: 37334379
- DOI: https://doi.org/10.3389/fimmu.2023.1178355
- Primary Citation of Related Structures:
7S4S, 7URQ, 7URS - PubMed Abstract:
SARS-CoV-2, the virus behind the COVID-19 pandemic, has changed over time to the extent that the current virus is substantially different from what originally led to the pandemic in 2019-2020. Viral variants have modified the severity and transmissibility of the disease and continue do so. How much of this change is due to viral fitness versus a response to immune pressure is hard to define. One class of antibodies that continues to afford some level of protection from emerging variants are those that closely overlap the binding site for angiotensin-converting enzyme 2 (ACE2) on the receptor binding domain (RBD). Some members of this class that were identified early in the course of the pandemic arose from the V H 3-53 germline gene (IGHV3-53*01) and had short heavy chain complementarity-determining region 3s (CDR H3s). Here, we describe the molecular basis of the SARS-CoV-2 RBD recognition by the anti-RBD monoclonal antibody CoV11 isolated early in the COVID-19 pandemic and show how its unique mode of binding the RBD determines its neutralization breadth. CoV11 utilizes a heavy chain V H 3-53 and a light chain V K 3-20 germline sequence to bind to the RBD. Two of CoV11's four heavy chain changes from the V H 3-53 germline sequence,
h r F W R H 1 28 e r C D R H 1 31
Organizational Affiliation:
Infectious Disease Division, Department of Medicine of Uniformed Services University of the Health Sciences, Bethesda, MD, United States.