Crystal structure of AO75L
Laugeri, M.E., Speciale, I., Gimeno, A., Lin, S., Poveda, A., Lowary, T., Van Etten, J.L., Barbero, J.J., De Castro, C., Tonetti, M., Rojas, A.L.To be published.
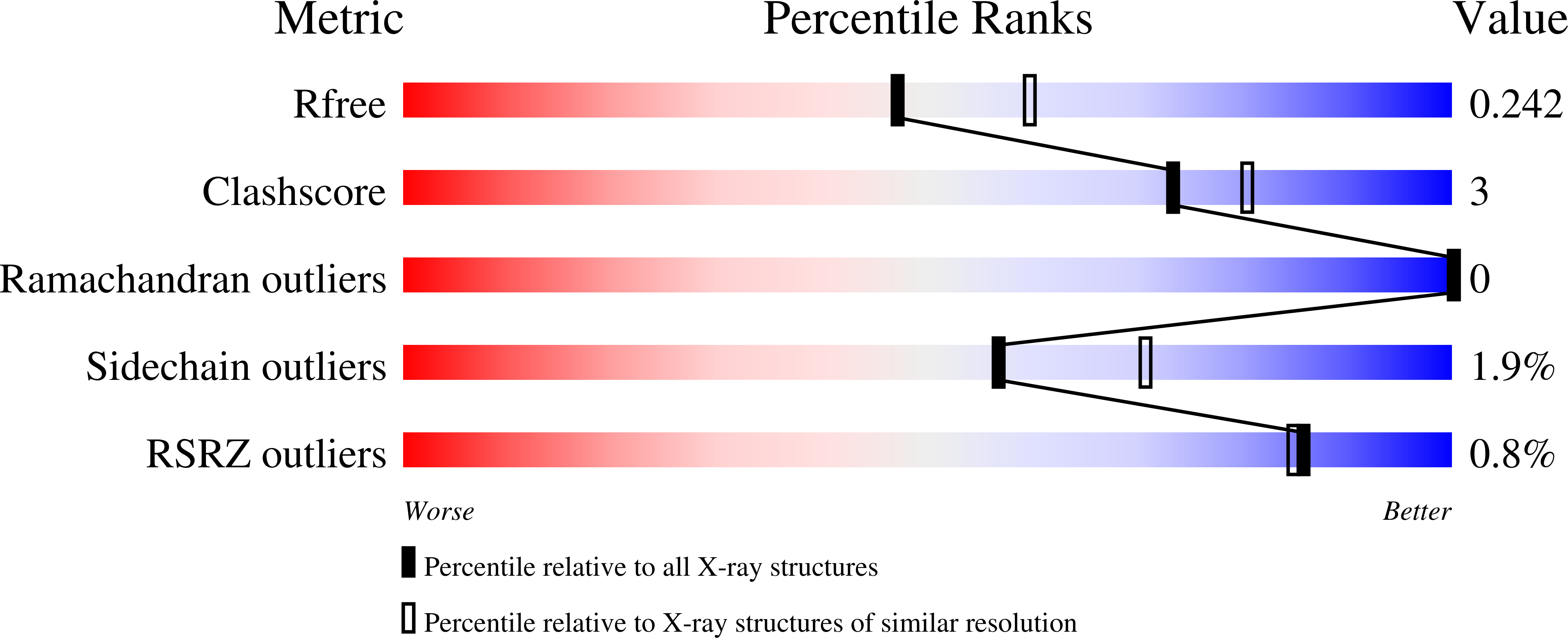
Experimental Data Snapshot
Biological assembly 1 assigned by authors.
Biological assembly 2 assigned by authors.
Biological assembly 3 assigned by authors.
Biological assembly 4 assigned by authors.
Biological assembly 5 assigned by authors.
Biological assembly 6 assigned by authors.
Biological assembly 7 assigned by authors.
Biological assembly 8 assigned by authors.
Macromolecule Content
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Exostosin | 285 | Paramecium bursaria Chlorella virus 1 | Mutation(s): 0 Gene Names: A075L | 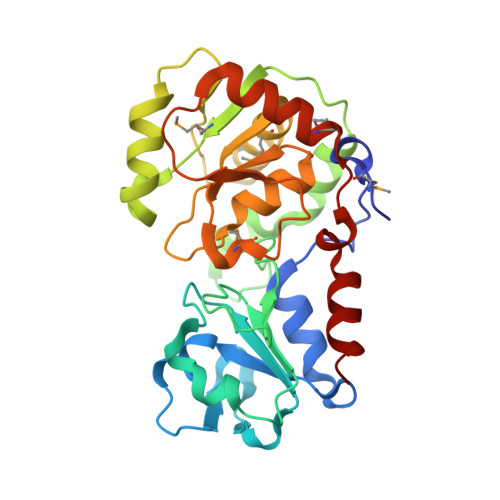 | |
UniProt | |||||
Find proteins for Q89410 (Paramecium bursaria Chlorella virus 1) Explore Q89410 Go to UniProtKB: Q89410 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q89410 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Modified Residues 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Type | Formula | 2D Diagram | Parent |
| MSE Query on MSE | A, B, C, D, E A, B, C, D, E, F, G, H | L-PEPTIDE LINKING | C5 H11 N O2 Se |  | MET |
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 86.708 | α = 90.46 |
| b = 90.772 | β = 108.6 |
| c = 91.676 | γ = 109.71 |
| Software Name | Purpose |
|---|---|
| XDS | data reduction |
| XSCALE | data scaling |
| REFMAC | refinement |
| PDB_EXTRACT | data extraction |
| PHENIX | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Spanish Ministry of Science, Innovation, and Universities | Spain | SEV20160644 |
RCSB PDB Core Operations are funded by the U.S. National Science Foundation (DBI-2321666), the US Department of Energy (DE-SC0019749), and the National Cancer Institute, National Institute of Allergy and Infectious Diseases, and National Institute of General Medical Sciences of the National Institutes of Health under grant R01GM157729.