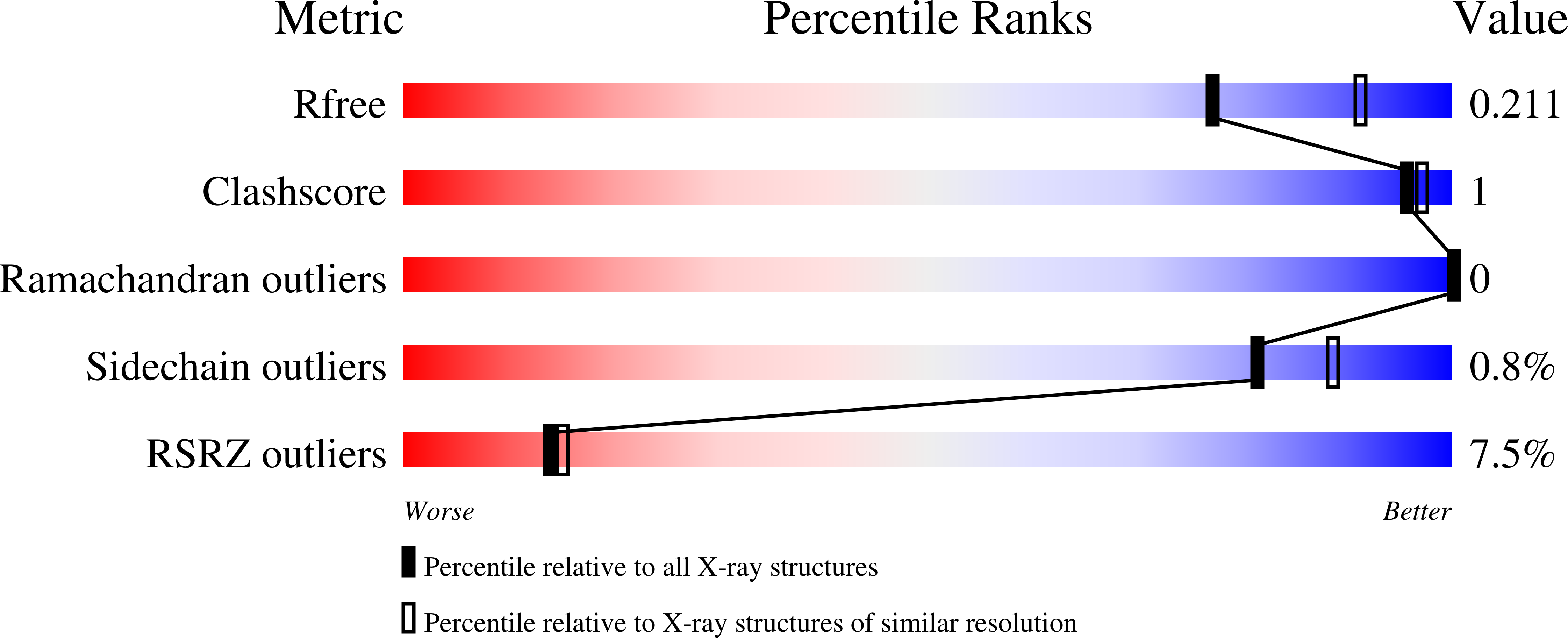
A metal-dependent conformational change provides a structural basis for the inhibition of CTP synthase by gemcitabine-5'-triphosphate.
McLeod, M.J., Tran, N., McCluskey, G.D., Gillis, T.D., Bearne, S.L., Holyoak, T.(2023) Protein Sci 32: e4648-e4648
- PubMed: 37106216
- DOI: https://doi.org/10.1002/pro.4648
- Primary Citation of Related Structures:
8FV6, 8FV7, 8FV8, 8FV9, 8FVA, 8FVB, 8FVC, 8FVD, 8FVE, 8SBR - PubMed Abstract:
CTP synthases (CTPS) catalyze the de novo production of CTP using UTP, ATP, and l-glutamine with the anticancer drug metabolite gemcitabine-5'-triphosphate (dF-dCTP) being one of its most potent nucleotide inhibitors. To delineate the structural origins of this inhibition, we solved the structures of Escherichia coli CTPS (ecCTPS) in complex with CTP (2.0 Å), 2'-ribo-F-dCTP (2.0 Å), 2'-arabino-F-CTP (2.4 Å), dF-dCTP (2.3 Å), dF-dCTP and ADP (2.1 Å), and dF-dCTP and ATP (2.1 Å). These structures revealed that the increased binding affinities observed for inhibitors bearing the 2'-F-arabino group (dF-dCTP and F-araCTP), relative to CTP and F-dCTP, arise from interactions between the inhibitor's fluorine atom exploiting a conserved hydrophobic pocket formed by F227 and an interdigitating loop from an adjacent subunit (Q114-V115-I116). Intriguingly, crystal structures of ecCTPS•dF-dCTP complexes in the presence of select monovalent and divalent cations demonstrated that the in crystallo tetrameric assembly of wild-type ecCTPS was induced into a conformation similar to inhibitory ecCTPS filaments solely through the binding of Na + -, Mg 2+ -, or Mn 2+ •dF-dCTP. However, in the presence of potassium, the dF-dCTP-bound structure is demetalated and in the low-affinity, non-filamentous conformation, like the conformation seen when bound to CTP and the other nucleotide analogues. Additionally, CTP can also induce the filament-competent conformation linked to high-affinity dF-dCTP binding in the presence of high concentrations of Mg 2+ . This metal-dependent, compacted CTP pocket conformation therefore furnishes the binding environment responsible for the tight binding of dF-dCTP and provides insights for further inhibitor design.
Organizational Affiliation:
Department of Biology, University of Waterloo, Waterloo, Ontario, Canada.