Drug-induced rotational movement of the ribosome is a key factor for read-through enhancement
Kolosova, O., Zgadzay, Y., Yusupov, M.To be published.
Experimental Data Snapshot
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 60S ribosomal protein L2-B | D [auth j], DC [auth AW] | 254 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A8H6F1X9 (Candida albicans) Explore A0A8H6F1X9 Go to UniProtKB: A0A8H6F1X9 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A8H6F1X9 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 5 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 60S ribosomal protein L3 | E [auth k], EC [auth AX] | 389 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for Q59LS1 (Candida albicans (strain SC5314 / ATCC MYA-2876)) Explore Q59LS1 Go to UniProtKB: Q59LS1 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q59LS1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 6 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 60S ribosomal protein L4-B | F [auth l], FC [auth AY] | 363 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A8H6F4H4 (Candida albicans) Explore A0A8H6F4H4 Go to UniProtKB: A0A8H6F4H4 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A8H6F4H4 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 7 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Uncharacterized protein CaJ7.0206 | G [auth m], GC [auth AZ] | 298 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for G1UAA6 (Candida albicans) Explore G1UAA6 Go to UniProtKB: G1UAA6 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | G1UAA6 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 8 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 60S ribosomal protein L6 | H [auth n], HC [auth BA] | 176 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for Q9P834 (Candida albicans) Explore Q9P834 Go to UniProtKB: Q9P834 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q9P834 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 9 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 60S ribosomal protein L7-A | I [auth o], IC [auth BB] | 241 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A8H6BVN2 (Candida albicans) Explore A0A8H6BVN2 Go to UniProtKB: A0A8H6BVN2 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A8H6BVN2 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 10 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 60S ribosomal protein L8 | J [auth p], JC [auth BC] | 262 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A8H6C337 (Candida albicans) Explore A0A8H6C337 Go to UniProtKB: A0A8H6C337 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A8H6C337 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 11 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 60S ribosomal protein L9-B | K [auth q], KC [auth BD] | 191 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A8H6C3N3 (Candida albicans) Explore A0A8H6C3N3 Go to UniProtKB: A0A8H6C3N3 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A8H6C3N3 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 12 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 60S ribosomal protein L10 | L [auth r], LC [auth BE] | 220 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for Q5AIB8 (Candida albicans (strain SC5314 / ATCC MYA-2876)) Explore Q5AIB8 Go to UniProtKB: Q5AIB8 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q5AIB8 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 13 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 60S ribosomal protein L11-B | M [auth s], MC [auth BF] | 174 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A8H6F1S5 (Candida albicans) Explore A0A8H6F1S5 Go to UniProtKB: A0A8H6F1S5 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A8H6F1S5 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 14 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 60S ribosomal protein L13 | N [auth t], NC [auth BG] | 202 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A8H6F5V7 (Candida albicans) Explore A0A8H6F5V7 Go to UniProtKB: A0A8H6F5V7 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A8H6F5V7 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 15 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 60S ribosomal protein L14-B | O [auth u], OC [auth BH] | 131 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A8H6F332 (Candida albicans) Explore A0A8H6F332 Go to UniProtKB: A0A8H6F332 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A8H6F332 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 16 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 60S ribosomal protein L15-A | P [auth v], PC [auth BI] | 204 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for Q5A6R1 (Candida albicans (strain SC5314 / ATCC MYA-2876)) Explore Q5A6R1 Go to UniProtKB: Q5A6R1 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q5A6R1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 17 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Ribosomal protein L13 | Q [auth w], QC [auth BJ] | 200 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A8H6BZ56 (Candida albicans) Explore A0A8H6BZ56 Go to UniProtKB: A0A8H6BZ56 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A8H6BZ56 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 18 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Ribosomal protein L22 | R [auth x], RC [auth BK] | 185 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A8H6BUG0 (Candida albicans) Explore A0A8H6BUG0 Go to UniProtKB: A0A8H6BUG0 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A8H6BUG0 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 19 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 60S ribosomal protein L18-A | S [auth y], SC [auth BL] | 186 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A8H6C4Y1 (Candida albicans) Explore A0A8H6C4Y1 Go to UniProtKB: A0A8H6C4Y1 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A8H6C4Y1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 20 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 60S ribosomal protein L19-A | T [auth z], TC [auth BM] | 190 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A1D8PK40 (Candida albicans (strain SC5314 / ATCC MYA-2876)) Explore A0A1D8PK40 Go to UniProtKB: A0A1D8PK40 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A1D8PK40 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 21 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 60S ribosomal protein L20 | U [auth 0], UC [auth BN] | 172 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A8H6BZJ3 (Candida albicans) Explore A0A8H6BZJ3 Go to UniProtKB: A0A8H6BZJ3 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A8H6BZJ3 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 22 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 60S ribosomal protein L21-A | V [auth 2], VC [auth BO] | 160 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A8H6F1C8 (Candida albicans) Explore A0A8H6F1C8 Go to UniProtKB: A0A8H6F1C8 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A8H6F1C8 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 23 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 60S ribosomal protein L22-B | W [auth 5], WC [auth BP] | 124 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A1D8PM41 (Candida albicans (strain SC5314 / ATCC MYA-2876)) Explore A0A1D8PM41 Go to UniProtKB: A0A1D8PM41 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A1D8PM41 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 24 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 60S ribosomal protein L23-A | X [auth 6], XC [auth BQ] | 137 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A8H6C444 (Candida albicans) Explore A0A8H6C444 Go to UniProtKB: A0A8H6C444 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A8H6C444 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 25 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 60S ribosomal protein L24-A | Y [auth 7], YC [auth BR] | 155 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A8H6C4U1 (Candida albicans) Explore A0A8H6C4U1 Go to UniProtKB: A0A8H6C4U1 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A8H6C4U1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 26 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 60S ribosomal protein L25 | Z [auth 8], ZC [auth BS] | 142 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A8H6F6A1 (Candida albicans) Explore A0A8H6F6A1 Go to UniProtKB: A0A8H6F6A1 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A8H6F6A1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 27 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Ribosomal protein L24 | AA [auth 9], AD [auth BT] | 127 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A8H6F3X2 (Candida albicans) Explore A0A8H6F3X2 Go to UniProtKB: A0A8H6F3X2 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A8H6F3X2 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 28 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 60S ribosomal protein L27 | BA [auth AA], BD [auth BU] | 136 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for Q9P843 (Candida albicans) Explore Q9P843 Go to UniProtKB: Q9P843 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q9P843 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 29 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 60S ribosomal protein L28 | CA [auth AB], CD [auth BV] | 149 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A8H6BVY8 (Candida albicans) Explore A0A8H6BVY8 Go to UniProtKB: A0A8H6BVY8 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A8H6BVY8 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 30 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 60S ribosomal protein L29 | DA [auth AC], DD [auth BW] | 63 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A8H6F0G8 (Candida albicans) Explore A0A8H6F0G8 Go to UniProtKB: A0A8H6F0G8 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A8H6F0G8 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 31 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 60S ribosomal protein L30 | EA [auth AD], ED [auth BX] | 106 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A1D8PM75 (Candida albicans (strain SC5314 / ATCC MYA-2876)) Explore A0A1D8PM75 Go to UniProtKB: A0A1D8PM75 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A1D8PM75 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 32 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 60S ribosomal protein L31-B | FA [auth AE], FD [auth BY] | 112 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A8H6F1M1 (Candida albicans) Explore A0A8H6F1M1 Go to UniProtKB: A0A8H6F1M1 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A8H6F1M1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 33 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 60S ribosomal protein L32 | GA [auth AF], GD [auth BZ] | 131 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for O94008 (Candida albicans) Explore O94008 Go to UniProtKB: O94008 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | O94008 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 34 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 60S ribosomal protein L33-A | HA [auth AG], HD [auth CA] | 107 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A8H6BW57 (Candida albicans) Explore A0A8H6BW57 Go to UniProtKB: A0A8H6BW57 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A8H6BW57 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 35 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 60S ribosomal protein L34-B | IA [auth AH], ID [auth CB] | 122 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A8H6F171 (Candida albicans) Explore A0A8H6F171 Go to UniProtKB: A0A8H6F171 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A8H6F171 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 36 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Ribosomal protein L29 | JA [auth AI], JD [auth CC] | 120 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A8H6F6B7 (Candida albicans) Explore A0A8H6F6B7 Go to UniProtKB: A0A8H6F6B7 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A8H6F6B7 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 37 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 60S ribosomal protein L36 | KA [auth AJ], KD [auth CD] | 99 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for P47834 (Candida albicans) Explore P47834 Go to UniProtKB: P47834 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P47834 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 38 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 60S ribosomal protein L37-B | LA [auth AK], LD [auth CE] | 90 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A1D8PF45 (Candida albicans (strain SC5314 / ATCC MYA-2876)) Explore A0A1D8PF45 Go to UniProtKB: A0A1D8PF45 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A1D8PF45 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 39 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 60S ribosomal protein L38 | MA [auth AL], MD [auth CF] | 78 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A1D8PG16 (Candida albicans (strain SC5314 / ATCC MYA-2876)) Explore A0A1D8PG16 Go to UniProtKB: A0A1D8PG16 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A1D8PG16 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 40 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 60S ribosomal protein L39 | NA [auth AM], ND [auth CG] | 51 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for Q96W55 (Candida albicans) Explore Q96W55 Go to UniProtKB: Q96W55 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q96W55 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 41 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 60S ribosomal protein L40-B | OA [auth AN], OD [auth CH] | 52 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A1D8PL68 (Candida albicans (strain SC5314 / ATCC MYA-2876)) Explore A0A1D8PL68 Go to UniProtKB: A0A1D8PL68 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A1D8PL68 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar proteins by: Sequence | 3D Structure
Entity ID: 42 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 60S ribosomal protein L41 | PA [auth AO], PD [auth CI] | 25 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for P9WER7 (Candida albicans (strain SC5314 / ATCC MYA-2876)) Explore P9WER7 Go to UniProtKB: P9WER7 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P9WER7 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 43 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 60S ribosomal protein L42-B | QA [auth AP], QD [auth CJ] | 106 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A8H6BR69 (Candida albicans) Explore A0A8H6BR69 Go to UniProtKB: A0A8H6BR69 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A8H6BR69 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 44 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 60S ribosomal protein L43-A | RA [auth AQ], RD [auth CK] | 92 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A1D8PP14 (Candida albicans (strain SC5314 / ATCC MYA-2876)) Explore A0A1D8PP14 Go to UniProtKB: A0A1D8PP14 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A1D8PP14 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 46 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 40S ribosomal protein S0 | TA [auth C], TD [auth CN] | 261 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A8H6C395 (Candida albicans) Explore A0A8H6C395 Go to UniProtKB: A0A8H6C395 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A8H6C395 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 47 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 40S ribosomal protein S1 | UA [auth D], UD [auth CO] | 256 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A8H6C387 (Candida albicans) Explore A0A8H6C387 Go to UniProtKB: A0A8H6C387 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A8H6C387 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 48 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Ribosomal protein S5 | VA [auth E], VD [auth CP] | 249 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A8H6BZ90 (Candida albicans) Explore A0A8H6BZ90 Go to UniProtKB: A0A8H6BZ90 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A8H6BZ90 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 49 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Ribosomal protein S3 | WA [auth F], WD [auth CQ] | 251 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A8H6BYS1 (Candida albicans) Explore A0A8H6BYS1 Go to UniProtKB: A0A8H6BYS1 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A8H6BYS1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 50 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 40S ribosomal protein S4 | XA [auth G], XD [auth CR] | 262 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A1D8PCI6 (Candida albicans (strain SC5314 / ATCC MYA-2876)) Explore A0A1D8PCI6 Go to UniProtKB: A0A1D8PCI6 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A1D8PCI6 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 51 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Ribosomal protein S7 | YA [auth H], YD [auth CS] | 225 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A8H6BS29 (Candida albicans) Explore A0A8H6BS29 Go to UniProtKB: A0A8H6BS29 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A8H6BS29 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 52 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 40S ribosomal protein S6 | ZA [auth I], ZD [auth CT] | 236 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A8H6BZ37 (Candida albicans) Explore A0A8H6BZ37 Go to UniProtKB: A0A8H6BZ37 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A8H6BZ37 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 53 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 40S ribosomal protein S7 | AB [auth J], AE [auth CU] | 186 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A8H6C655 (Candida albicans) Explore A0A8H6C655 Go to UniProtKB: A0A8H6C655 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A8H6C655 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 54 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 40S ribosomal protein S8 | BB [auth K], BE [auth CV] | 206 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A8H6F0S0 (Candida albicans) Explore A0A8H6F0S0 Go to UniProtKB: A0A8H6F0S0 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A8H6F0S0 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 55 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Ribosomal protein S4 | CB [auth L], CE [auth CW] | 189 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A8H6F1X2 (Candida albicans) Explore A0A8H6F1X2 Go to UniProtKB: A0A8H6F1X2 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A8H6F1X2 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 56 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 40S ribosomal protein S10-A | DB [auth M], DE [auth CX] | 118 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A8H6BWQ8 (Candida albicans) Explore A0A8H6BWQ8 Go to UniProtKB: A0A8H6BWQ8 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A8H6BWQ8 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 57 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 40S ribosomal protein S11A | EB [auth N], EE [auth CY] | 155 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A1D8PN83 (Candida albicans (strain SC5314 / ATCC MYA-2876)) Explore A0A1D8PN83 Go to UniProtKB: A0A1D8PN83 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A1D8PN83 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 58 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 40S ribosomal protein S12 | FB [auth O], FE [auth CZ] | 143 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A8H6C3A1 (Candida albicans) Explore A0A8H6C3A1 Go to UniProtKB: A0A8H6C3A1 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A8H6C3A1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 59 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 40S ribosomal protein S13 | GB [auth P], GE [auth DA] | 151 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A8H6F4G0 (Candida albicans) Explore A0A8H6F4G0 Go to UniProtKB: A0A8H6F4G0 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A8H6F4G0 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 60 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 40S ribosomal protein S14-A | HB [auth Q], HE [auth DB] | 132 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A8H6F0V4 (Candida albicans) Explore A0A8H6F0V4 Go to UniProtKB: A0A8H6F0V4 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A8H6F0V4 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 61 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 40S ribosomal protein S15 | IB [auth R], IE [auth DC] | 142 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A1D8PK22 (Candida albicans (strain SC5314 / ATCC MYA-2876)) Explore A0A1D8PK22 Go to UniProtKB: A0A1D8PK22 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A1D8PK22 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 62 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 40S ribosomal protein S16 | JB [auth S], JE [auth DD] | 142 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for O94017 (Candida albicans) Explore O94017 Go to UniProtKB: O94017 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | O94017 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 63 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 40S ribosomal protein S17-B | KB [auth T], KE [auth DE] | 137 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A8H6BTR7 (Candida albicans) Explore A0A8H6BTR7 Go to UniProtKB: A0A8H6BTR7 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A8H6BTR7 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 64 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 40S ribosomal protein S18-B | LB [auth U], LE [auth DF] | 145 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A8H6F3Y8 (Candida albicans) Explore A0A8H6F3Y8 Go to UniProtKB: A0A8H6F3Y8 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A8H6F3Y8 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 65 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 40S ribosomal protein S19-A | MB [auth V], ME [auth DG] | 145 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A8H6C5L4 (Candida albicans) Explore A0A8H6C5L4 Go to UniProtKB: A0A8H6C5L4 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A8H6C5L4 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 66 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Ribosomal protein S10 | NB [auth W], NE [auth DH] | 119 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A8H6BYJ9 (Candida albicans) Explore A0A8H6BYJ9 Go to UniProtKB: A0A8H6BYJ9 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A8H6BYJ9 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 67 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 40S ribosomal protein S21 | OB [auth X], OE [auth DI] | 87 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for Q9P844 (Candida albicans) Explore Q9P844 Go to UniProtKB: Q9P844 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q9P844 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 68 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 40S ribosomal protein S22-A | PB [auth Y], PE [auth DJ] | 130 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A8H6F0N4 (Candida albicans) Explore A0A8H6F0N4 Go to UniProtKB: A0A8H6F0N4 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A8H6F0N4 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 69 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Ribosomal protein S23 (S12) | QB [auth Z], QE [auth DK] | 145 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A8H6F0V0 (Candida albicans) Explore A0A8H6F0V0 Go to UniProtKB: A0A8H6F0V0 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A8H6F0V0 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 70 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 40S ribosomal protein S24 | RB [auth a], RE [auth DL] | 135 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A8H6C3K1 (Candida albicans) Explore A0A8H6C3K1 Go to UniProtKB: A0A8H6C3K1 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A8H6C3K1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 71 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 40S ribosomal protein S25 | SB [auth b], SE [auth DM] | 105 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A8H6EZQ8 (Candida albicans) Explore A0A8H6EZQ8 Go to UniProtKB: A0A8H6EZQ8 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A8H6EZQ8 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 72 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 40S ribosomal protein S26 | TB [auth c], TE [auth DN] | 119 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A8H6BTS1 (Candida albicans) Explore A0A8H6BTS1 Go to UniProtKB: A0A8H6BTS1 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A8H6BTS1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 73 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 40S ribosomal protein S27 | UB [auth d], UE [auth DO] | 82 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A8H6BWE6 (Candida albicans) Explore A0A8H6BWE6 Go to UniProtKB: A0A8H6BWE6 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A8H6BWE6 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 74 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 40S ribosomal protein S28-B | VB [auth e], VE [auth DP] | 67 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A8H6C0A5 (Candida albicans) Explore A0A8H6C0A5 Go to UniProtKB: A0A8H6C0A5 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A8H6C0A5 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 75 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 40S ribosomal protein S29A | WB [auth f], WE [auth DQ] | 56 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A1D8PTR4 (Candida albicans (strain SC5314 / ATCC MYA-2876)) Explore A0A1D8PTR4 Go to UniProtKB: A0A1D8PTR4 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A1D8PTR4 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 76 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 40S ribosomal protein S30 | XB [auth g], XE [auth DR] | 63 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A1D8PSK2 (Candida albicans (strain SC5314 / ATCC MYA-2876)) Explore A0A1D8PSK2 Go to UniProtKB: A0A1D8PSK2 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A1D8PSK2 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 77 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Ubiquitin-40S ribosomal protein S31 fusion protein | YB [auth h] | 193 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for Q5A109 (Candida albicans (strain SC5314 / ATCC MYA-2876)) Explore Q5A109 Go to UniProtKB: Q5A109 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q5A109 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 78 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Guanine nucleotide-binding protein subunit beta-like protein | ZB [auth AR] | 317 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A8H6C3E7 (Candida albicans) Explore A0A8H6C3E7 Go to UniProtKB: A0A8H6C3E7 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A8H6C3E7 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 79 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 60S ribosomal protein CAALFM_C304810CA | YE [auth i], ZE [auth CL] | 267 | Candida albicans | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A1D8PK71 (Candida albicans (strain SC5314 / ATCC MYA-2876)) Explore A0A1D8PK71 Go to UniProtKB: A0A1D8PK71 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A1D8PK71 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| 25S | A [auth 1], AC [auth AS] | 3,359 | Candida albicans | 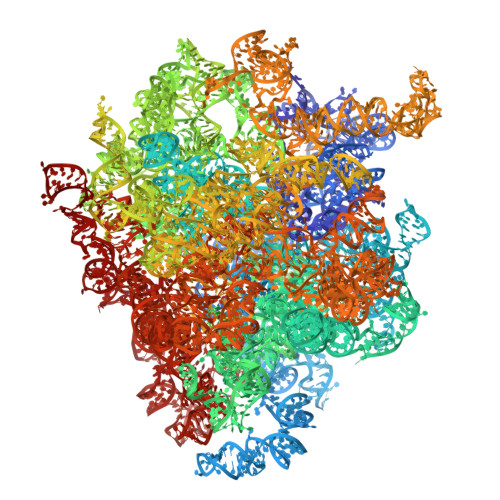 | |
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| 5S | B [auth 3], BC [auth AT] | 121 | Candida albicans | 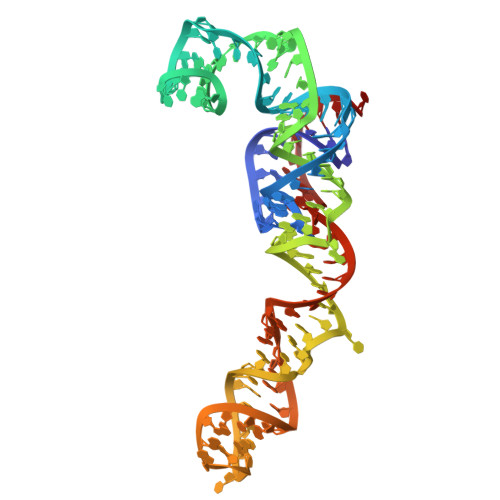 | |
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| 5.8S | C [auth 4], CC [auth AU] | 158 | Candida albicans |  | |
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 45 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| 18S | SA [auth B], SD [auth CM] | 1,787 | Candida albicans | 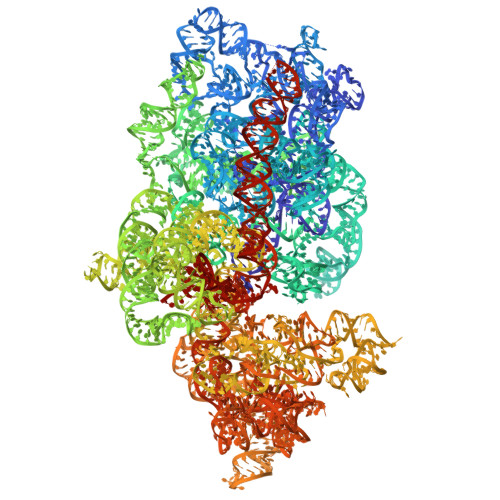 | |
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 4 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| 3K5 Query on 3K5 | AF [auth 1] | 3-O-acetyl-2-O-(3-O-acetyl-6-deoxy-beta-D-glucopyranosyl)-6-deoxy-1-O-{[(2R,2'S,3a'R,4''S,5''R,6'S,7a'S)-5''-methyl-4''-{[(2E)-3-phenylprop-2-enoyl]oxy}decahydrodispiro[oxirane-2,3'-[1]benzofuran-2',2''-pyran]-6'-yl]carbonyl}-beta-D-glucopyranose C40 H52 O17 VOTNXJVGRXZYOA-XFJWYURVSA-N |  | ||
| PAR (Subject of Investigation/LOI) Query on PAR | ABA [auth AS] AG [auth 1] BBA [auth AS] BF [auth 1] BG [auth 1] | PAROMOMYCIN C23 H45 N5 O14 UOZODPSAJZTQNH-LSWIJEOBSA-N |  | ||
| ZN Query on ZN | EUA [auth DN] FOA [auth CB] FUA [auth DO] GOA [auth CE] GUA [auth DQ] | ZINC ION Zn PTFCDOFLOPIGGS-UHFFFAOYSA-N |  | ||
| MG Query on MG | AAA [auth B] ACA [auth AS] ADA [auth AS] AEA [auth AS] AFA [auth AS] | MAGNESIUM ION Mg JLVVSXFLKOJNIY-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 297.3 | α = 90 |
| b = 292.54 | β = 99.37 |
| c = 442.98 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| XDS | data reduction |
| XSCALE | data scaling |
| PHENIX | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Foundation for Medical Research (France) | France | DEQ20181039600 |
| Foundation for Medical Research (France) | France | DBF20160635745 |
| Foundation for Medical Research (France) | France | FDT202204014886 |
| Russian Science Foundation | Russian Federation | 20-65-47031 |