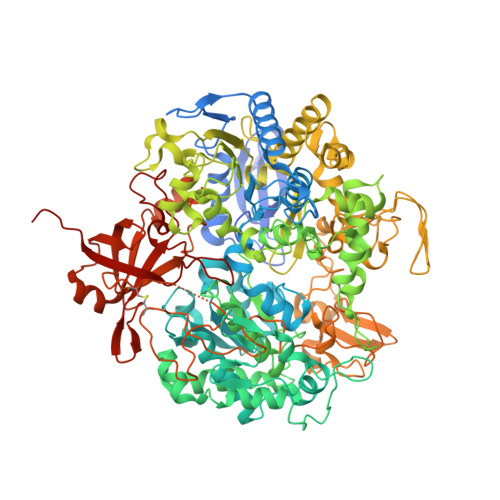
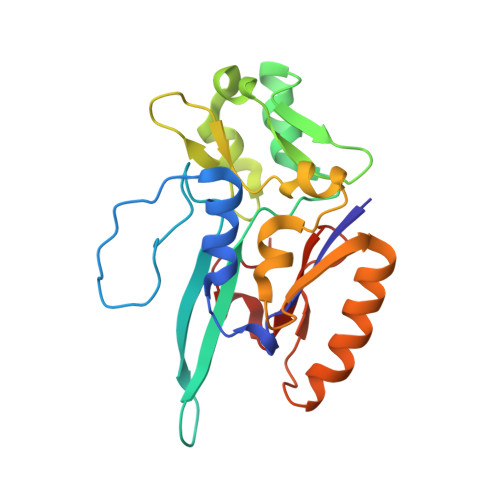
Substrate-dependent oxidative inactivation of a W-dependent formate dehydrogenase involving selenocysteine displacement.
Vilela-Alves, G., Manuel, R.R., Viegas, A., Carpentier, P., Biaso, F., Guigliarelli, B., Pereira, I.A.C., Romao, M.J., Mota, C.(2024) Chem Sci 15: 13090-13101
- PubMed: 39148770
- DOI: https://doi.org/10.1039/d4sc02394c
- Primary Citation of Related Structures:
8RC8, 8RC9, 8RCA, 8RCB, 8RCC - PubMed Abstract:
Metal-dependent formate dehydrogenases are very promising targets for enzyme optimization and design of bio-inspired catalysts for CO 2 reduction, towards innovative strategies for climate change mitigation. For effective application of these enzymes, the catalytic mechanism must be better understood, and the molecular determinants clarified. Despite numerous studies, several doubts persist, namely regarding the role played by the possible dissociation of the SeCys ligand from the Mo/W active site. Additionally, the oxygen sensitivity of these enzymes must also be understood as it poses an important obstacle for biotechnological applications. This work presents a combined biochemical, spectroscopic, and structural characterization of Desulfovibrio vulgaris FdhAB ( Dv FdhAB) when exposed to oxygen in the presence of a substrate (formate or CO 2 ). This study reveals that O 2 inactivation is promoted by the presence of either substrate and involves forming a different species in the active site, captured in the crystal structures, where the SeCys ligand is displaced from tungsten coordination and replaced by a dioxygen or peroxide molecule. This form was reproducibly obtained and supports the conclusion that, although W- Dv FdhAB can catalyse the oxidation of formate in the presence of oxygen for some minutes, it gets irreversibly inactivated after prolonged O 2 exposure in the presence of either substrate.
Organizational Affiliation:
Associate Laboratory i4HB - Institute for Health and Bioeconomy, NOVA School of Science and Technology, Universidade NOVA de Lisboa 2829-516 Caparica Portugal [email protected] [email protected].