Structural mechanism of the ligand-activated heterodimeric AHR-ARNT complex
Diao, X., Shang, Q., Guo, M., Huang, Y., Zhang, M., Chen, X., Liang, Y., Sun, X., Zhuang, J., Liu, S., Vogel, C.F.A., Rastinejad, F., Wu, D.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Aryl hydrocarbon receptor nuclear translocator | 382 | Homo sapiens | Mutation(s): 0 Gene Names: ARNT | 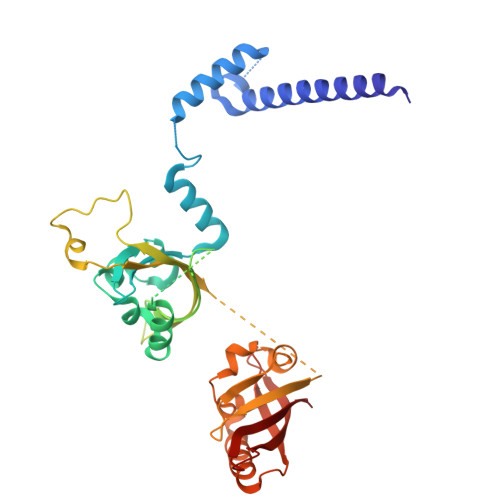 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P27540 (Homo sapiens) Explore P27540 Go to UniProtKB: P27540 | |||||
PHAROS: P27540 GTEx: ENSG00000143437 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P27540 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Aryl hydrocarbon receptor | 395 | Sus scrofa | Mutation(s): 0 Gene Names: AHR | 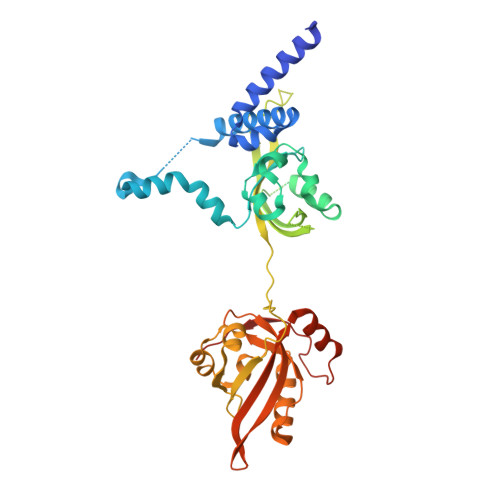 | |
UniProt | |||||
Find proteins for I3LF82 (Sus scrofa) Explore I3LF82 Go to UniProtKB: I3LF82 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | I3LF82 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar nucleic acids by: Sequence | 3D Structure
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| DNAF | 21 | Homo sapiens |  | ||
Sequence AnnotationsExpand | |||||
| |||||
Find similar nucleic acids by: Sequence | 3D Structure
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| DNAR | 21 | Homo sapiens |  | ||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| A1LWK (Subject of Investigation/LOI) Query on A1LWK | F [auth B] | 2-(3-oxidanyl-1H-indol-2-yl)indol-3-one C16 H10 N2 O2 QQILFGKZUJYXGS-UHFFFAOYSA-N |  | ||
| DTT Query on DTT | E [auth A] | 2,3-DIHYDROXY-1,4-DITHIOBUTANE C4 H10 O2 S2 VHJLVAABSRFDPM-IMJSIDKUSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 68.016 | α = 90 |
| b = 98.837 | β = 90.41 |
| c = 80.307 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| HKL-3000 | data scaling |
| HKL-3000 | data reduction |
| PHENIX | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Natural Science Foundation of China (NSFC) | China | 82373876 |