Structural and virologic mechanism of the emergence of resistance to M pro inhibitors in SARS-CoV-2.
Hattori, S.I., Bulut, H., Hayashi, H., Kishimoto, N., Takamune, N., Hasegawa, K., Furusawa, Y., Yamayoshi, S., Murayama, K., Tamamura, H., Li, M., Wlodawer, A., Kawaoka, Y., Misumi, S., Mitsuya, H.(2024) Proc Natl Acad Sci U S A 121: e2404175121-e2404175121
- PubMed: 39236245
- DOI: https://doi.org/10.1073/pnas.2404175121
- Primary Citation of Related Structures:
9ARQ, 9ARS, 9ART, 9AVQ - PubMed Abstract:
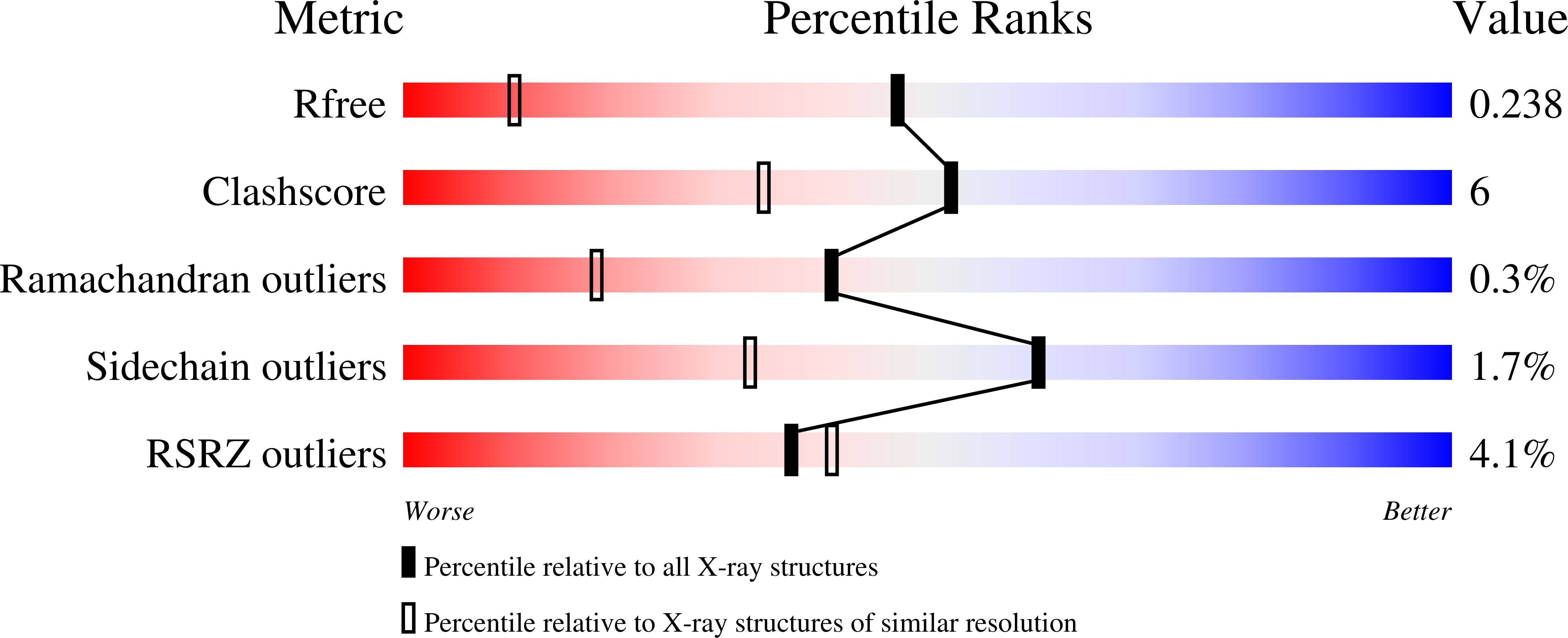
We generated SARS-CoV-2 variants resistant to three SARS-CoV-2 main protease (M pro ) inhibitors (nirmatrelvir, TKB245, and 5h), by propagating the ancestral SARS-CoV-2 WK521 WT in VeroE6 TMPRSS2 cells with increasing concentrations of each inhibitor and examined their structural and virologic profiles. A predominant E166V-carrying variant (SARS-CoV-2 WK521 E166V ), which emerged when passaged with nirmatrelvir and TKB245, proved to be resistant to the two inhibitors. A recombinant SARS-CoV-2 E166V was resistant to nirmatrelvir and TKB245, but sensitive to 5h. X-ray structural study showed that the dimerization of M pro was severely hindered by E166V substitution due to the disruption of the presumed dimerization-initiating Ser1'-Glu166 interactions. TKB245 stayed bound to M pro E166V , whereas nirmatrelvir failed. Native mass spectrometry confirmed that nirmatrelvir and TKB245 promoted the dimerization of M pro , and compromised the enzymatic activity; the Ki values of recombinant M pro E166V for nirmatrelvir and TKB245 were 117±3 and 17.1±1.9 µM, respectively, indicating that TKB245 has a greater (by a factor of 6.8) binding affinity to M pro E166V than nirmatrelvir. SARS-CoV-2 WK521 WT selected with 5h acquired A191T substitution in M pro (SARS-CoV-2 WK521 A191T ) and better replicated in the presence of 5h, than SARS-CoV-2 WK521 WT . However, no significant enzymatic or structural changes in M pro A191T were observed. The replicability of SARS-CoV-2 WK521 E166V proved to be compromised compared to SARS-CoV-2 WK521 WT but predominated over SARS-CoV-2 WK521 WT in the presence of nirmatrelvir. The replicability of SARS-CoV-2 WK521 A191T surpassed that of SARS-CoV-2 WK521 WT in the absence of 5h, confirming that A191T confers enhanced viral fitness. The present data should shed light on the understanding of the mechanism of SARS-CoV-2's drug resistance acquisition and the development of resistance-repellant COVID-19 therapeutics.
Organizational Affiliation:
Department of Refractory Viral Diseases, National Center for Global Health and Medicine Research Institute, Tokyo 162-8655, Japan.