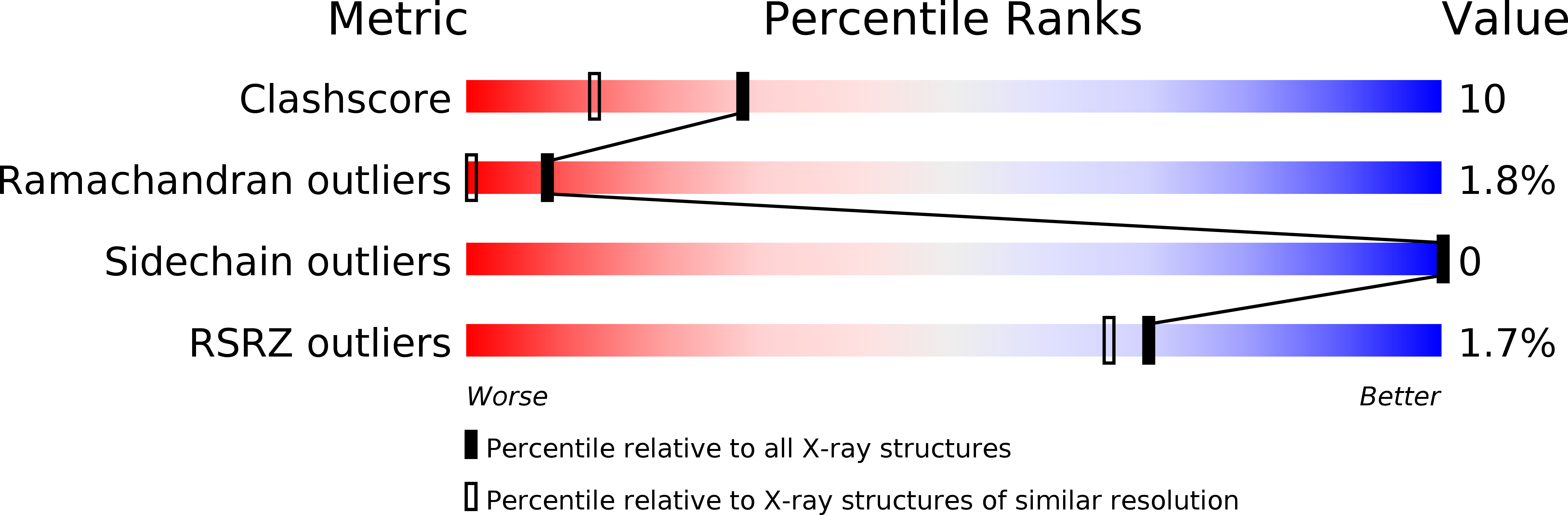Structure of bovine pancreatic trypsin inhibitor at 125 K definition of carboxyl-terminal residues Gly57 and Ala58.
Parkin, S., Rupp, B., Hope, H.(1996) Acta Crystallogr D Biol Crystallogr 52: 18-29
- PubMed: 15299722
- DOI: https://doi.org/10.1107/S0907444995008675
- Primary Citation of Related Structures:
1BPI - PubMed Abstract:
The structure of bovine pancreatic trypsin inhibitor has been refined to a resolution of 1.1 A against data collected at 125 K. The space group of the form II crystal is P2(1)2(1)2(1) with a = 75.39(3), b = 22.581(7), c = 28.606 (9) A (cf. a = 74.1, b = 23.4, c = 28.9 A at room temperature). The structure was refined by restrained least-squares minimization of summation operator w(F (o)(2)- F (c)(2))(2) with the SHELXL93 program. As the model improved, water molecules were included and exceptionally clear electron density was found for two residues, Gly57 and Ala58, that had been largely obscured at room temperature. The side chains of residues Glu7 and Arg53 were modelled over two positions with refined occupancy factors. The final model contains 145.6 water molecules distributed over 167 sites, and a single phosphate group disordered over two sites. The root-mean-square discrepancy between Calpha atoms in residues Arg1-Gly56 at room and low temperatures is 0.4 A. A comparison of models refined with anisotropic and isotropic thermal parameters revealed that there were no significant differences in atomic positions. The final weighted R-factor on F(2) (wR(2)) for data in the range 10-1.1 A was 35.9% for the anisotropic model and 40.9% for the isotropic model. Conventional R-factors based on F for F > 4sigma(F) were 12.2 and 14.6%, respectively, corresponding to 16.1 and 18.7% on all data. These large R-factor differences were not reflected in values of R(free), which were not significantly different at 21.5(5) and 21.8(4)%, respectively. These results, along with the relatively straightforward nature of the refinement, clearly highlight the benefits of low-temperature data collection.
Organizational Affiliation:
Department of Chemistry, University of California, Davis 95616, USA.