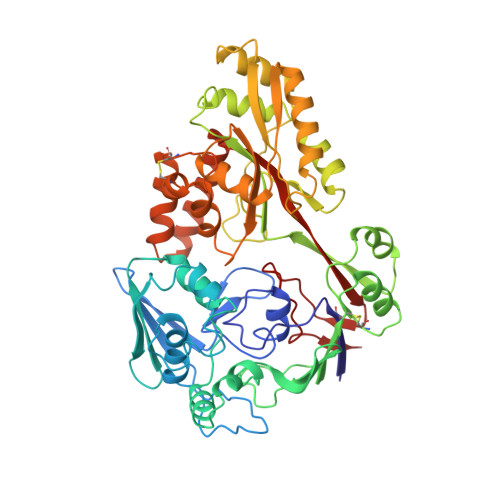
2 A resolution structure of DppA, a periplasmic dipeptide transport/chemosensory receptor.
Nickitenko, A.V., Trakhanov, S., Quiocho, F.A.(1995) Biochemistry 34: 16585-16595
- PubMed: 8527431
- DOI: https://doi.org/10.1021/bi00051a006
- Primary Citation of Related Structures:
1DPE - PubMed Abstract:
The family of about 50 periplasmic binding proteins, which exhibit diverse specificity (e.g., carbohydrates, amino acids, dipeptides, oligopeptides, oxyanions, metals, and vitamins) and range in size from 20 to 58 kDa, is a gold mine for an atomic-level investigation of structure and molecular recognition. These proteins serve as initial receptors for active transport systems or permeases. About six of these proteins, including the dipeptide-binding protein (DppA), are also primary receptors for chemotaxis. The structure of the unbound form of DppA (M(r) = 57,400) has been determined and refined to an R-factor of 0.169 to 2 A resolution. DppA consists of two distinct domains (I and II) connected by two "hinge" segments which form part of the base of the wide groove between the two domains. The relative orientation of the two domains gives the protein a pearlike shape, with domain I and domain II forming the larger and smaller apical ends, respectively. From the tip to the rounded bottom measures about 85 A, and the widest diameter is about 60 A. Domain I, which consists of two integrated subdomains, is folded from two separate polypeptide segments from the amino- and carboxyl-terminal ends. The more compact domain II is formed from the intervening segment. Comparison of the dipeptide-binding protein structure with that of the bound form of the similar oligopeptide-binding protein [Tame, J. R. H., Murshudov, G. N., Dodson, E. J., Neil, T. K., Dodson, G. G., Higgins, C. F., & Wilkinson, A. J. (1994) Science 264, 1578-1581] reveals the major features that differentiate the ligand specificity of the two proteins and describe the large hinge bending (about 55 degrees) between the two domains.
Organizational Affiliation:
Howard Hughes Medical Institute, Baylor College of Medicine, Houston, Texas 77030, USA.