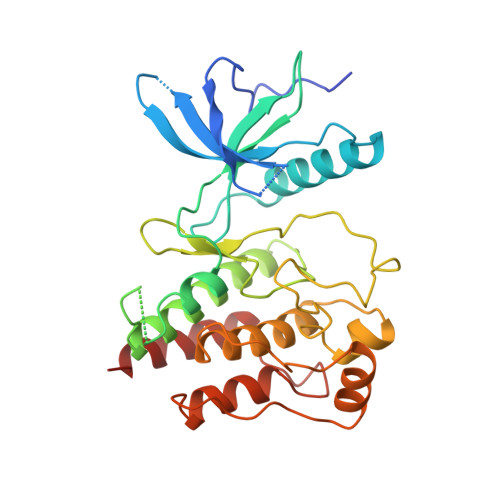
Structure of the FGF receptor tyrosine kinase domain reveals a novel autoinhibitory mechanism.
Mohammadi, M., Schlessinger, J., Hubbard, S.R.(1996) Cell 86: 577-587
- PubMed: 8752212
- DOI: https://doi.org/10.1016/s0092-8674(00)80131-2
- Primary Citation of Related Structures:
1FGK - PubMed Abstract:
The crystal structure of the tyrosine kinase domain of fibroblast growth factor receptor 1 (FGFR1K) has been determined in its unliganded form to 2.0 angstroms resolution and in complex with with an ATP analog to 2.3 angstrosms A resolution. Several features distinguish the structure of FGFR1K from that of the tyrosine kinase domain of the insulin receptor. Residues in the activation loop of FGFR1K appear to interfere with substrate peptide binding but not with ATP binding, revealing a second and perhaps more general autoinhibitory mechanism for receptor tyrosine kinases. In addition, a dimeric form of FGFR1K observed in the crystal structure may provide insights into the molecular mechanisms by which FGF receptors are activated. Finally, the structure provides a basis for rationalizing the effects of kinase mutations in FGF receptors that lead to developmental disorders in nematodes and humans.
Organizational Affiliation:
Department of Pharmacology, New York University Medical Center, New York 10016, USA.