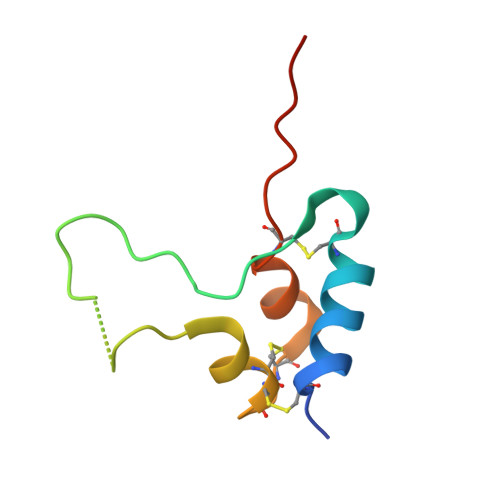
Structural Origins of the Functional Divergence of Human Insulin-Like Growth Factor-I and Insulin
Brzozowski, A.M., Dodson, E.J., Dodson, G.G., Murshudov, G., Verma, C., Turkenburg, J.P., De Bree, F.M., Dauter, Z.(2002) Biochemistry 41: 9389
- PubMed: 12135360
- DOI: https://doi.org/10.1021/bi020084j
- Primary Citation of Related Structures:
1GZR, 1GZY, 1GZZ, 1H02 - PubMed Abstract:
Human insulin-like growth factors I and II (hIGF-I, hIGF-II) are potent stimulators of cell and growth processes. They display high sequence similarity to both the A and B chains of insulin but contain an additional connecting C-domain, which reflects their secretion without specific packaging or precursor conversion. IGFs also have an extension at the C-terminus known as the D-domain. This paper describes four homologous hIGF-1 structures, obtained from crystals grown in the presence of the detergent SB12, which reveal additional detail in the C- and D-domains. Two different detergent binding modes observed in the crystals may reflect different hIGF-I biological properties such as the interaction with IGF binding proteins and self-aggregation. While the helical core of hIGF-I is very similar to that in insulin, there are distinct differences in the region of hIGF-I corresponding to the insulin B chain C-terminus, residues B25-B30. In hIGF-I, these residues (24-29) and the following C-domain form an extensive loop protruding 20 A from the core, which results in a substantially different conformation for the receptor binding epitope in hIGF-I compared to insulin. One notable feature of the structures presented here is demonstration of peptide-bond cleavage between Ser35 and Arg36 resulting in an apparent gap between residues 35 and 39. The equivalent region of proinsulin is involved in hormone processing demanding a reassessment of the structural integrity of hIGF-I in relation to its biological function.
Organizational Affiliation:
Structural Biology Laboratory, Chemistry Department, University of York, Heslington, York YO10 5DD, United Kingdom. [email protected]