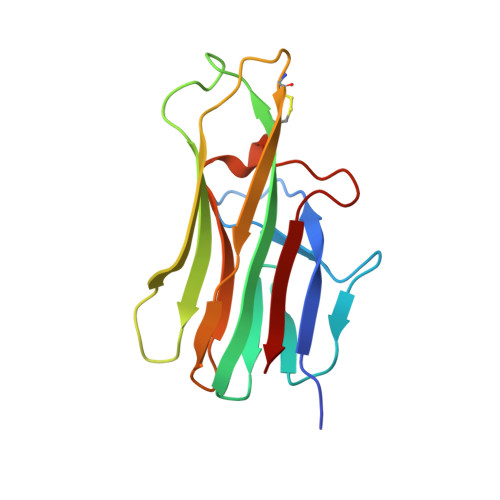
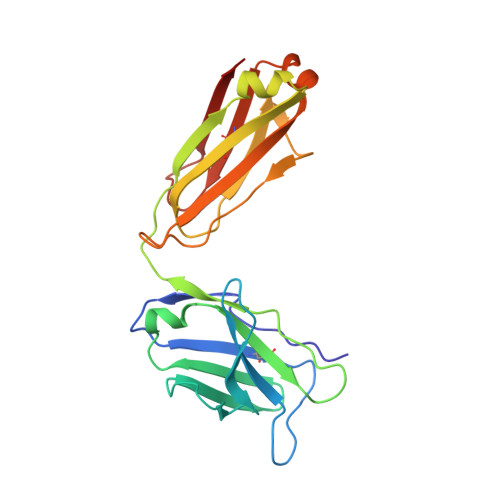
Structure of CD40 ligand in complex with the Fab fragment of a neutralizing humanized antibody.
Karpusas, M., Lucci, J., Ferrant, J., Benjamin, C., Taylor, F.R., Strauch, K., Garber, E., Hsu, Y.M.(2001) Structure 9: 321-329
- PubMed: 11525169
- DOI: https://doi.org/10.1016/s0969-2126(01)00590-1
- Primary Citation of Related Structures:
1I9R - PubMed Abstract:
CD40 ligand (CD40L or CD154), a member of the tumor necrosis factor (TNF) family, plays a critical role in both humoral and cellular immune responses and has been implicated in biological pathways involving epithelial cells, fibroblasts, and platelets. Such a pathway is T cell-mediated B cell activation, a process that occurs through the interaction of CD40L with CD40 receptor expressed on B cells. It results in various B cell responses, including immunoglobulin isotype switching and B cell differentiation and proliferation. These responses can be inhibited by the monoclonal antibody 5c8, which binds with high affinity to CD40L. To understand the structural basis of the inhibition, we determined the crystal structure of the complex of the extracellular domain of CD40L and the Fab fragment of humanized 5c8 antibody. The structure shows that the complex has the shape of a three-bladed propeller with three Fab fragments bound symmetrically to a CD40L homotrimer. To further study the nature of the antibody-antigen interface, we assessed the ability of 23 site-directed mutants of CD40L to bind to 5c8 and CD40 and analyzed the results in the context of the crystal structure. Finally, we observed via confocal microscopy that 5c8 binding to CD40L on the cell surface results in the formation of patches of clustered complexes. The structure reveals that 5c8 neutralizes CD40L function by sterically blocking CD40 binding. The antigenic epitope is localized in a region of the surface that is likely to be structurally perturbed as a result of genetic mutations that cause hyper-IgM syndrome. The symmetric trimeric arrangement of the Fab fragments in the complex results in a geometry that facilitates the formation of large clusters of complexes on the cell surface.
Organizational Affiliation:
Biogen, Inc, Cambridge, Massachusetts 02142, USA. [email protected].