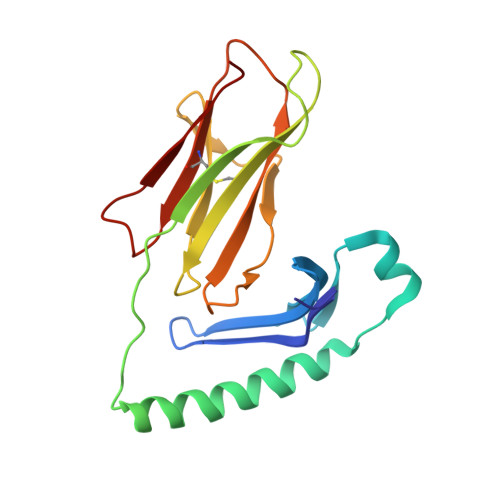
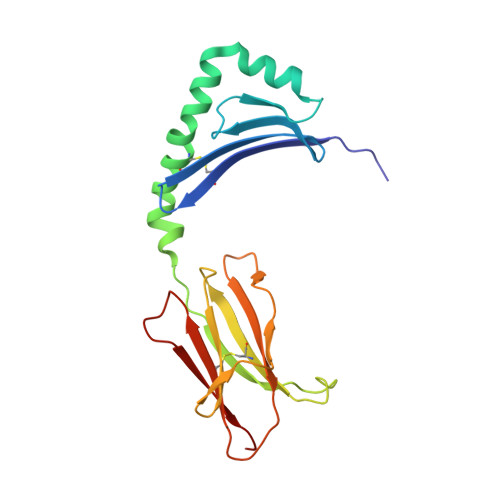
Structure of a human insulin peptide-HLA-DQ8 complex and susceptibility to type 1 diabetes.
Lee, K.H., Wucherpfennig, K.W., Wiley, D.C.(2001) Nat Immunol 2: 501-507
- PubMed: 11376336
- DOI: https://doi.org/10.1038/88694
- Primary Citation of Related Structures:
1JK8 - PubMed Abstract:
The class II major histocompatibility complex (MHC) glycoproteins HLA-DQ8 and HLA-DQ2 in humans and I-A(g7) in nonobese diabetic (NOD) mice are the major risk factors for increased susceptibility to type 1 diabetes. Using X-ray crystallography, we have determined the three-dimensional structure of DQ8 complexed with an immunodominant peptide from insulin. The similarity of the DQ8, DQ2 and I-A(g7) peptide-binding pockets suggests that diabetes is caused by the same antigen-presentation event(s) in humans and NOD mice. Correlating type 1 diabetes epidemiology and MHC sequences with the DQ8 structure suggests that other structural features of the P9 pocket in addition to position 57 contribute to susceptibility to type 1 diabetes.
Organizational Affiliation:
Laboratory of Molecular Medicine, The Department of Medicine, The Children's Hospital, 320 Longwood Avenue, Boston, MA 02115, USA.