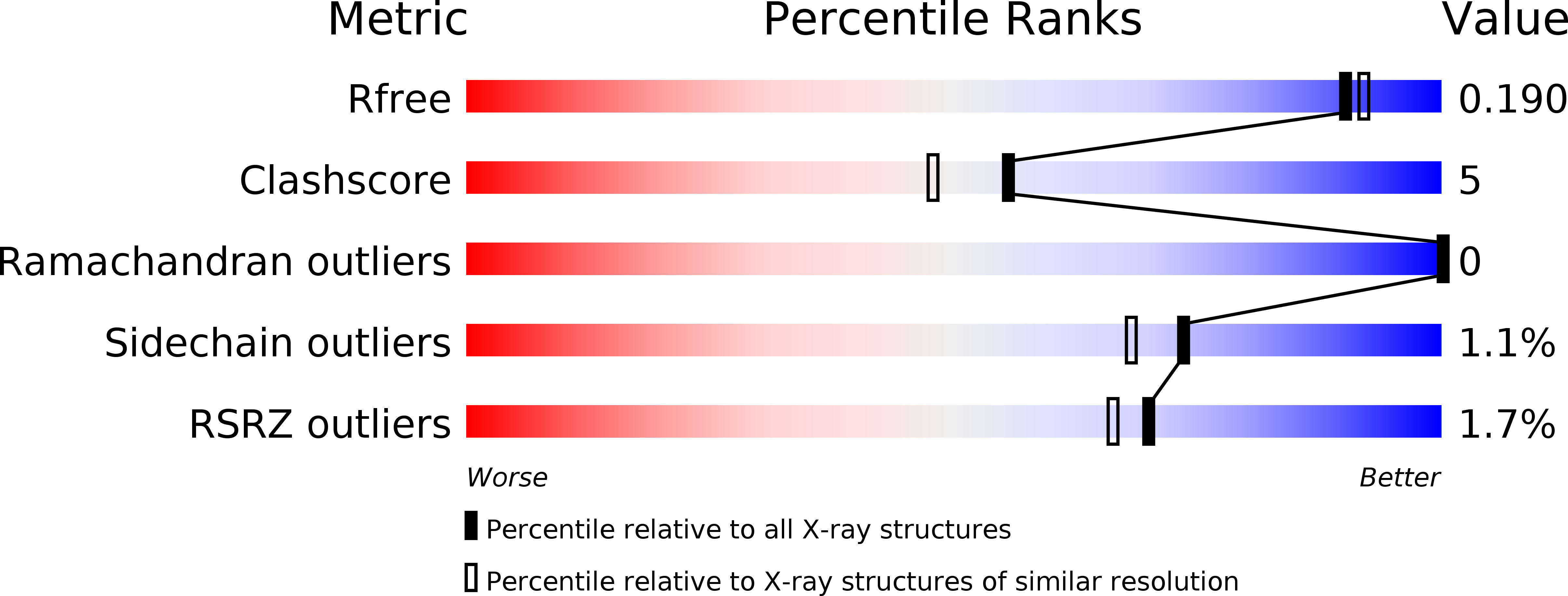
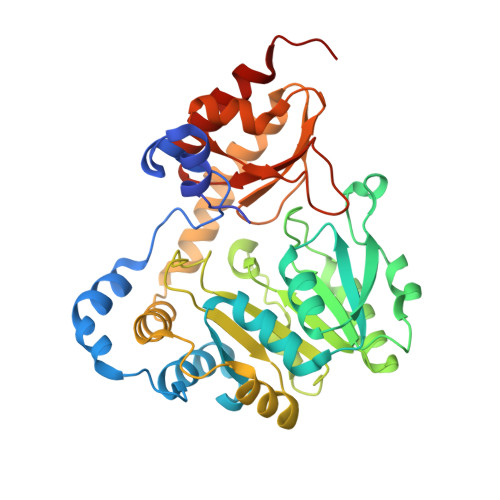
Three-dimensional structure of the L-threonine-O-3-phosphate decarboxylase (CobD) enzyme from Salmonella enterica.
Cheong, C.G., Bauer, C.B., Brushaber, K.R., Escalante-Semerena, J.C., Rayment, I.(2002) Biochemistry 41: 4798-4808
- PubMed: 11939774
- DOI: https://doi.org/10.1021/bi012111w
- Primary Citation of Related Structures:
1LKC - PubMed Abstract:
The three-dimensional structure of the pyridoxal 5'-phosphate (PLP)-dependent L-threonine-O-3-phosphate decarboxylase (CobD) from Salmonella enterica is described here. This enzyme is responsible for synthesizing (R)-1-amino-2-propanol phosphate which is the precursor for the linkage between the nucleotide loop and the corrin ring in cobalamin. The molecule is a molecular dimer where each subunit consists of a large and small domain. Overall the protein is very similar to the members of the family of aspartate aminotransferases. Indeed, the arrangement of the ligands surrounding the cofactor and putative substrate binding site are remarkably close to that observed in histidinol phosphate aminotransferase, which suggests that this latter enzyme might have been its progenitor. The only significant differences in structure occur at the N-terminus, which is approximately 12 residues shorter in CobD and does not form the same type of interdomain interaction common to other aminotransferases. CobD is unusual since within the aspartate aminotransferase subfamily of PLP-dependent enzymes the chemical transformations are substantially conserved, where the only exceptions are 1-aminocyclopropane-1-carboxylate synthase and CobD. Although there are a large number of PLP-dependent amino acid decarboxylases, these are generally larger and structurally distinct from the members of the aspartate aminotransferase subfamily of enzymes. The structure of CobD suggests that the chemical fate of the external aldimine can be redirected by modifications at the N-terminus of the protein. This study provides insight into the evolutionary history of the cobalamin biosynthetic pathway and raises the question of why most PLP-dependent decarboxylases are considerably larger enzymes.
Organizational Affiliation:
Department of Biochemistry, University of Wisconsin, Madison, Wisconsin 53706, USA.