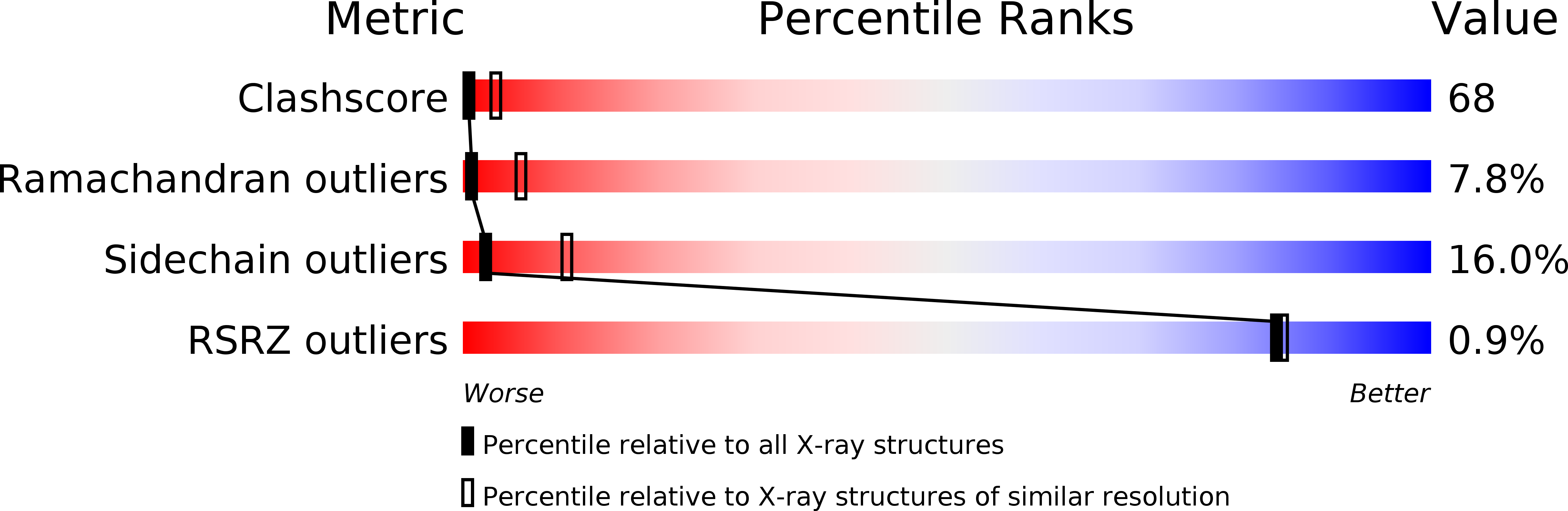The structure of a functional unit from the wall of a gastropod hemocyanin offers a possible mechanism for cooperativity
Perbandt, M., Guthoehrlein, E.W., Rypniewski, W., Idakieva, K., Stoeva, S., Voelter, W., Genov, N., Betzel, C.(2003) Biochemistry 42: 6341-6346
- PubMed: 12767214
- DOI: https://doi.org/10.1021/bi020672x
- Primary Citation of Related Structures:
1LNL - PubMed Abstract:
Structure-function relationships in a molluscan hemocyanin have been investigated by determining the crystal structure of the Rapana thomasiana (gastropod) hemocyanin functional unit RtH2e in deoxygenated form at 3.38 A resolution. This is the first X-ray structure of an unit from the wall of the molluscan hemocyanin cylinder. The crystal structure of RtH2e demonstrates molecular self-assembly of six identical molecules forming a regular hexameric cylinder. This suggests how the functional units are ordered in the wall of the native molluscan hemocyanins. The molecular arrangement is stabilized by specific protomer-to-protomer interactions, which are probably typical for the functional units building the wall of the cylinders. A molecular mechanism for cooperative dioxygen binding in molluscan hemocyanins is proposed on the basis of the molecular interactions between the protomers. In particular, the deoxygenated RtH2e structure reveals a tunnel leading from two opposite sides of the molecule to the active site. The tunnel represents a possible entrance pathway for dioxygen molecules. No such tunnels have been observed in the crystal structure of the oxy-Odg, a functional unit from the Octopus dofleini (cephalopod) hemocyanin in oxygenated form.
Organizational Affiliation:
Institute of Biochemistry and Molecular Biology I, University Hospital Hamburg-Eppendorf, c/o DESY, Building 22a, Notkestrasse 85, 22603 Hamburg, Germany.