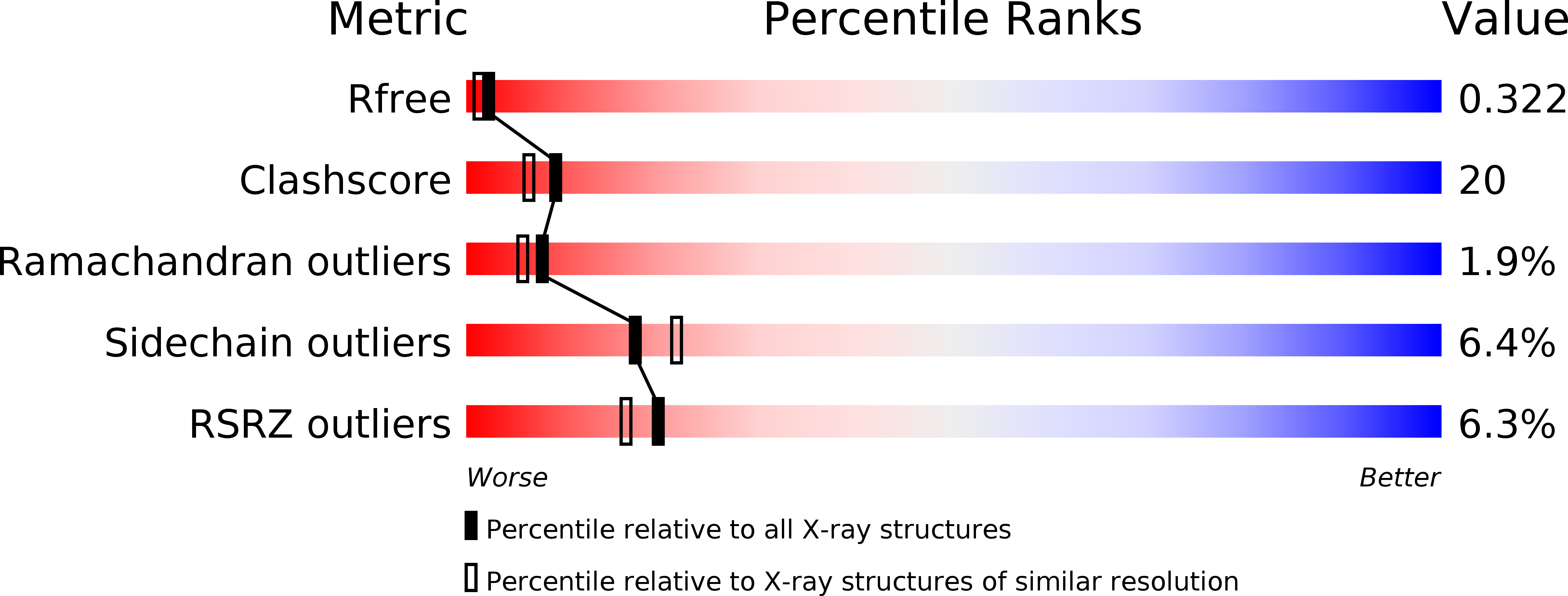
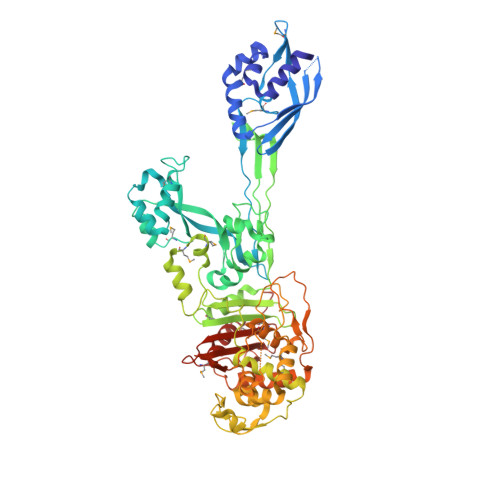
Structural basis for the beta lactam resistance of PBP2a from methicillin-resistant Staphylococcus aureus.
Lim, D., Strynadka, N.C.(2002) Nat Struct Biol 9: 870-876
- PubMed: 12389036
- DOI: https://doi.org/10.1038/nsb858
- Primary Citation of Related Structures:
1MWR, 1MWS, 1MWT, 1MWU, 1VQQ - PubMed Abstract:
The multiple antibiotic resistance of methicillin-resistant strains of Staphylococcus aureus (MRSA) has become a major clinical problem worldwide. The key determinant of the broad-spectrum beta-lactam resistance in MRSA strains is the penicillin-binding protein 2a (PBP2a). Because of its low affinity for beta-lactams, PBP2a provides transpeptidase activity to allow cell wall synthesis at beta-lactam concentrations that inhibit the beta-lactam-sensitive PBPs normally produced by S. aureus. The crystal structure of a soluble derivative of PBP2a has been determined to 1.8 A resolution and provides the highest resolution structure for a high molecular mass PBP. Additionally, structures of the acyl-PBP complexes of PBP2a with nitrocefin, penicillin G and methicillin allow, for the first time, a comparison of an apo and acylated resistant PBP. An analysis of the PBP2a active site in these forms reveals the structural basis of its resistance and identifies features in newly developed beta-lactams that are likely important for high affinity binding.
Organizational Affiliation:
2146 Health Sciences Mall, Department of Biochemistry and Molecular Biology, University of British Columbia, Vancouver, British Columbia V6T 1Z3, Canada.