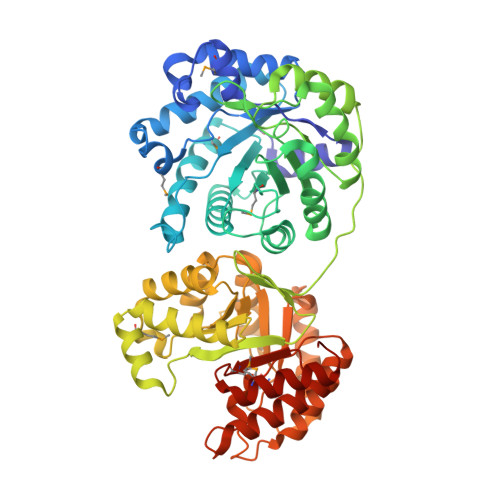
Structures of the N-terminal modules imply large domain motions during catalysis by methionine synthase.
Evans, J.C., Huddler, D.P., Hilgers, M.T., Romanchuk, G., Matthews, R.G., Ludwig, M.L.(2004) Proc Natl Acad Sci U S A 101: 3729-3736
- PubMed: 14752199
- DOI: https://doi.org/10.1073/pnas.0308082100
- Primary Citation of Related Structures:
1Q7M, 1Q7Q, 1Q7Z, 1Q85, 1Q8A, 1Q8J - PubMed Abstract:
B(12)-dependent methionine synthase (MetH) is a large modular enzyme that utilizes the cobalamin cofactor as a methyl donor or acceptor in three separate reactions. Each methyl transfer occurs at a different substrate-binding domain and requires a different arrangement of modules. In the catalytic cycle, the cobalamin-binding domain carries methylcobalamin to the homocysteine (Hcy) domain to form methionine and returns cob(I)alamin to the folate (Fol) domain for remethylation by methyltetrahydrofolate (CH(3)-H(4)folate). Here, we describe crystal structures of a fragment of MetH from Thermotoga maritima comprising the domains that bind Hcy and CH(3)-H(4)folate. These substrate-binding domains are (beta alpha)(8) barrels packed tightly against one another with their barrel axes perpendicular. The properties of the domain interface suggest that the two barrels remain associated during catalysis. The Hcy and CH(3)-H(4)folate substrates are bound at the C termini of their respective barrels in orientations that position them for reaction with cobalamin, but the two active sites are separated by approximately 50 A. To complete the catalytic cycle, the cobalamin-binding domain must travel back and forth between these distant active sites.
Organizational Affiliation:
Department of Biological Chemistry and Biophysics Research Division, University of Michigan, 930 North University Avenue, Ann Arbor, MI 48109-1055, USA.