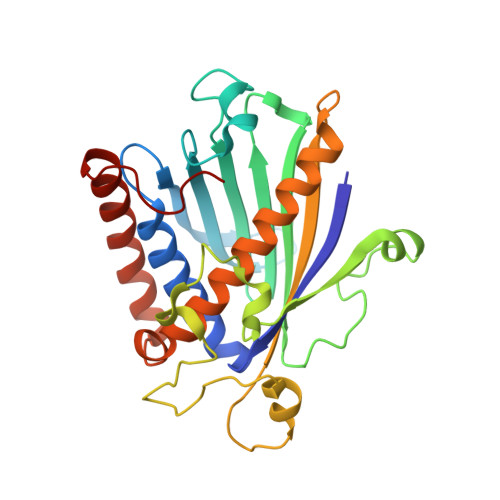
STRUCTURE OF A MULTIFUNCTIONAL PROTEIN. MAMMALIAN PHOSPHATIDYLINOSITOL TRANSFER PROTEIN COMPLEXED WITH PHOSPHATIDYLCHOLINE
Yoder, M.D., Thomas, L.M., Tremblay, J.M., Oliver, R.L., Yarbrough, L.R., Helmkamp Jr., G.M.(2001) J Biol Chem 276: 9246-9252
- PubMed: 11104777
- DOI: https://doi.org/10.1074/jbc.M010131200
- Primary Citation of Related Structures:
1T27 - PubMed Abstract:
Eukaryotic phosphatidylinositol transfer protein is a ubiquitous multifunctional protein that transports phospholipids between membrane surfaces and participates in cellular phospholipid metabolism during signal transduction and vesicular trafficking. The three-dimensional structure of the alpha-isoform of rat phosphatidylinositol transfer protein complexed with one molecule of phosphatidylcholine, one of its physiological ligands, has been determined to 2.2 A resolution by x-ray diffraction techniques. A single beta-sheet and several long alpha-helices define an enclosed internal cavity in which a single molecule of the phospholipid is accommodated with its polar head group in the center of the protein and fatty acyl chains projected toward the surface. Other structural features suggest mechanisms by which cytosolic phosphatidylinositol transfer protein interacts with membranes for lipid exchange and associates with a variety of lipid and protein kinases.
Organizational Affiliation:
Division of Cell Biology and Biophysics, School of Biological Sciences, University of Missouri-Kansas City, Kansas City, Missouri 64110-2499 , USA. [email protected]