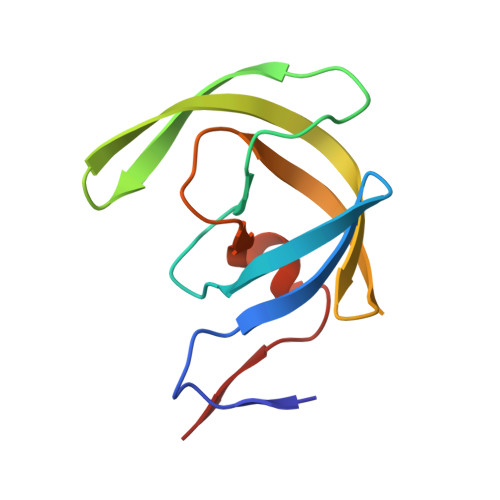
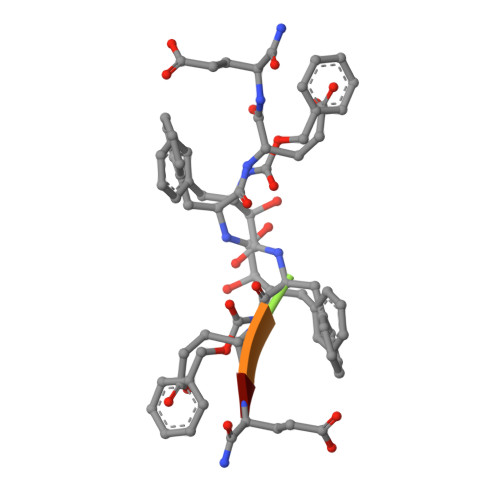
Inhibitor binding at the protein interface in crystals of a HIV-1 protease complex.
Brynda, J., Rezacova, P., Fabry, M., Horejsi, M., Stouracova, R., Soucek, M., Hradilek, M., Konvalinka, J., Sedlacek, J.(2004) Acta Crystallogr D Biol Crystallogr 60: 1943-1948
- PubMed: 15502300
- DOI: https://doi.org/10.1107/S0907444904021572
- Primary Citation of Related Structures:
1U8G - PubMed Abstract:
Depending on the excess of ligand used for complex formation, the HIV-1 protease complexed with a novel phenylnorstatine inhibitor forms crystals of either hexagonal (P6(1)) or orthorhombic (P2(1)2(1)2(1)) symmetry. The orthorhombic form shows an unusual complexity of crystal packing: in addition to one inhibitor molecule that is bound to the enzyme active site, the second inhibitor molecule is bound as an outer ligand at the protein interface. Binding of the outer ligand apparently increases the crystal-quality parameters so that the diffraction data allow solution of the structure of the complex at 1.03 A, the best resolution reported to date. The outer ligand interacts with all four surrounding HIV-1 protease molecules and has a bent conformation owing to its accommodation in the intermolecular space. The parameters of the solved structures of the orthorhombic and hexagonal forms are compared.
Organizational Affiliation:
Institute of Molecular Genetics, Academy of Sciences of the Czech Republic, Flemingovo nám. 2, 16637 Praha 6, Czech Republic. [email protected]