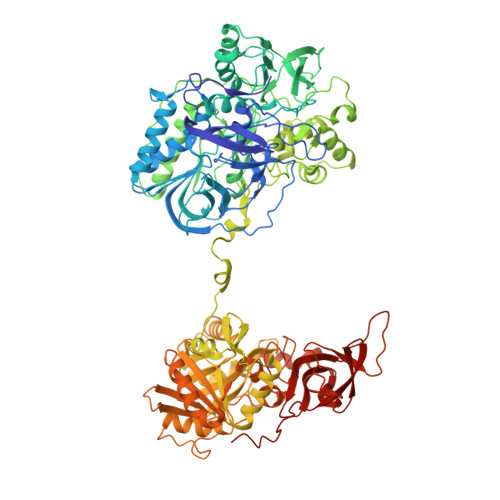
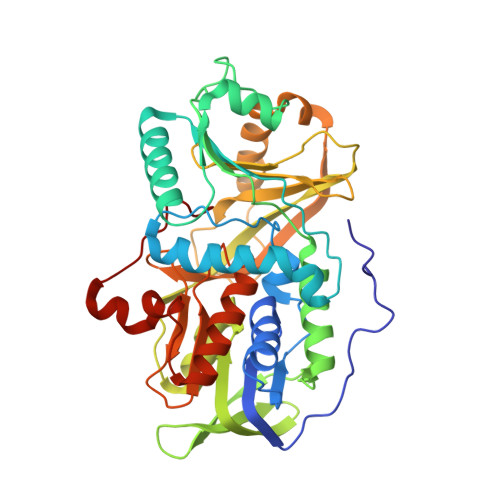
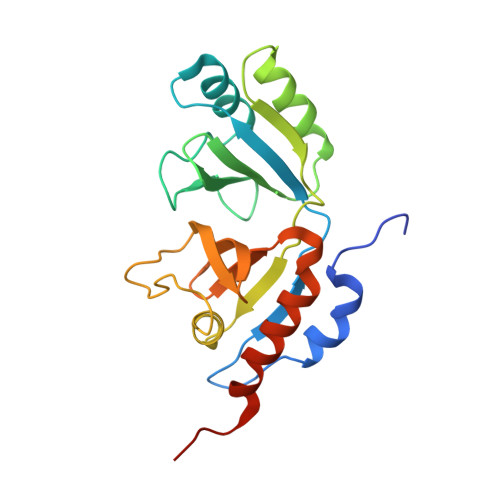
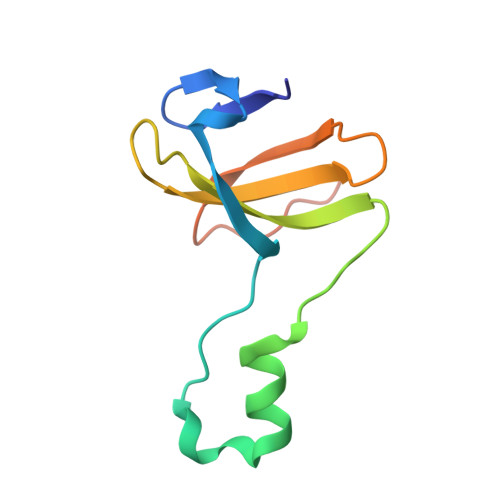
Crystal structure of heterotetrameric sarcosine oxidase from Corynebacterium sp. U-96
Ida, K., Moriguchi, T., Suzuki, H.(2005) Biochem Biophys Res Commun 333: 359-366
- PubMed: 15946648
- DOI: https://doi.org/10.1016/j.bbrc.2005.05.116
- Primary Citation of Related Structures:
1VRQ, 1X31 - PubMed Abstract:
Sarcosine oxidase from Corynebacterium sp. U-96 is a heterotetrameric enzyme. Here we report the crystal structures of the enzyme in complex with dimethylglycine and folinic acid. The alpha subunit is composed of two domains, contains NAD(+), and binds folinic acid. The beta subunit contains dimethylglycine, FAD, and FMN, and these flavins are approximately 10A apart. The gamma subunit is in contact with two domains of alpha subunit and has possibly a folate-binding structure. The delta subunit contains a single atom of zinc and has a Cys(3)His zinc finger structure. Based on the structures determined and on the previous works, the structure-function relationship on the heterotetrameric sarcosine oxidase is discussed.
Organizational Affiliation:
Department of Biosciences, School of Science, Kitasato University, 1-15-1 Kitasato, Sagamihara, Kanagawa 228-8555, Japan. [email protected]