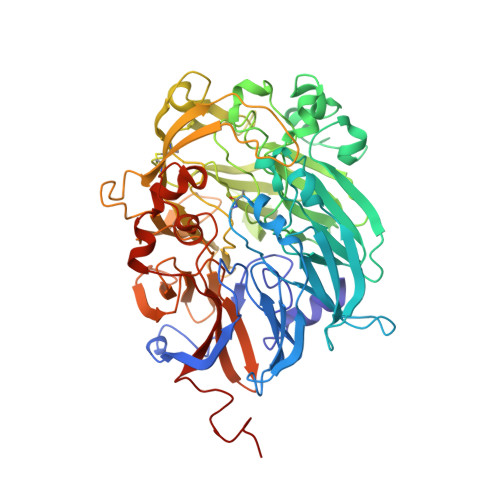
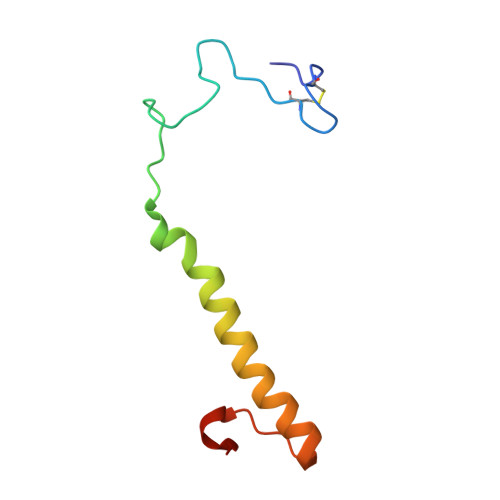
The Atomic Resolution Structure of Methanol Dehydrogenase from Methylobacterium Extorquens
Williams, P.A., Coates, L., Mohammed, F., Gill, R., Erskine, P.T., Wood, S.P., Anthony, C., Cooper, J.B.(2005) Acta Crystallogr D Biol Crystallogr 61: 75
- PubMed: 15608378
- DOI: https://doi.org/10.1107/S0907444904026964
- Primary Citation of Related Structures:
1W6S - PubMed Abstract:
The crystal structure of methanol dehydrogenase (MDH) from Methylobacterium extorquens has been refined without stereochemical restraints at a resolution of 1.2 A. The high-resolution data have defined the conformation of the tricyclic pyrroloquinoline quinone (PQQ) cofactor ring as entirely planar. The detailed definition of the active-site geometry has shown many features that are similar to the quinohaemo-protein alcohol dehydrogenases from Comamonas testosteroni and Pseudomonas putida, both of which possess MDH-like and cytochrome c-like domains. Conserved features between the two types of PQQ-containing enzyme suggest a common pathway for electron transfer between MDH and its physiological electron acceptor cytochrome cL. A pathway for proton transfer from the active site to the bulk solvent is also suggested.
Organizational Affiliation:
School of Biological Sciences, University of Southampton, Bassett Crescent East, Southampton SO16 7PX, England.