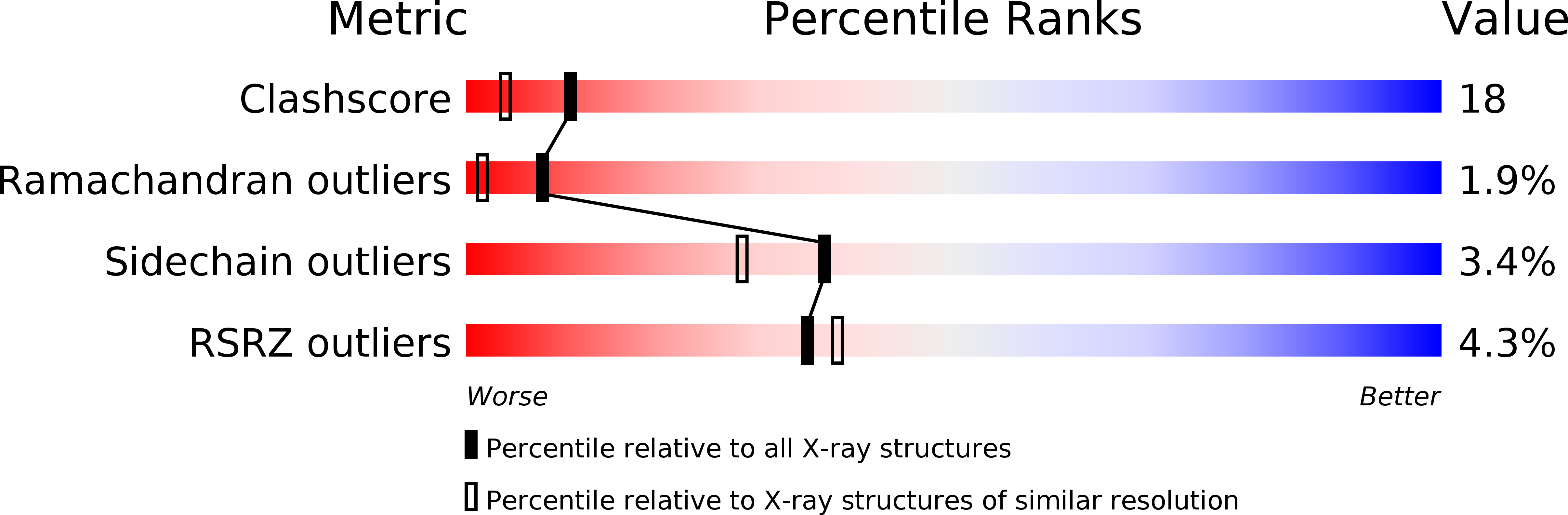Crystal Structure of a Polymeric Immunoglobulin Binding Fragment of the Human Polymeric Immunoglobulin Receptor
Hamburger, A.E., West Jr., A.P., Bjorkman, P.J.(2004) Structure 12: 1925-1935
- PubMed: 15530357
- DOI: https://doi.org/10.1016/j.str.2004.09.006
- Primary Citation of Related Structures:
1XED - PubMed Abstract:
The polymeric immunoglobulin receptor (pIgR) is a type I transmembrane protein that delivers dimeric IgA (dIgA) and pentameric IgM to mucosal secretions. Here, we report the 1.9 A resolution X-ray crystal structure of the N-terminal domain of human pIgR, which binds dIgA in the absence of other pIgR domains with an equilibrium dissociation constant of 300 nM. The structure of pIgR domain 1 reveals a folding topology similar to immunoglobulin variable domains, but with differences in the counterparts of the complementarity determining regions (CDRs), including a helical turn in CDR1 and a CDR3 loop that points away from the other CDRs. The unusual CDR3 loop position prevents dimerization analogous to the pairing of antibody variable heavy and variable light domains. The pIgR domain 1 structure allows interpretation of previous mutagenesis results and structure-based comparisons between pIgR and other IgA receptors.
Organizational Affiliation:
Division of Biology 114-96, California Institute of Technology, Pasadena, CA 91125, USA.