CO-trapping site in heme oxygenase revealed by photolysis of its co-bound heme complex: mechanism of escaping from product inhibition
Sugishima, M., Sakamoto, H., Noguchi, M., Fukuyama, K.(2004) J Mol Biol 341: 7-13
- PubMed: 15312758
- DOI: https://doi.org/10.1016/j.jmb.2004.05.048
- Primary Citation of Related Structures:
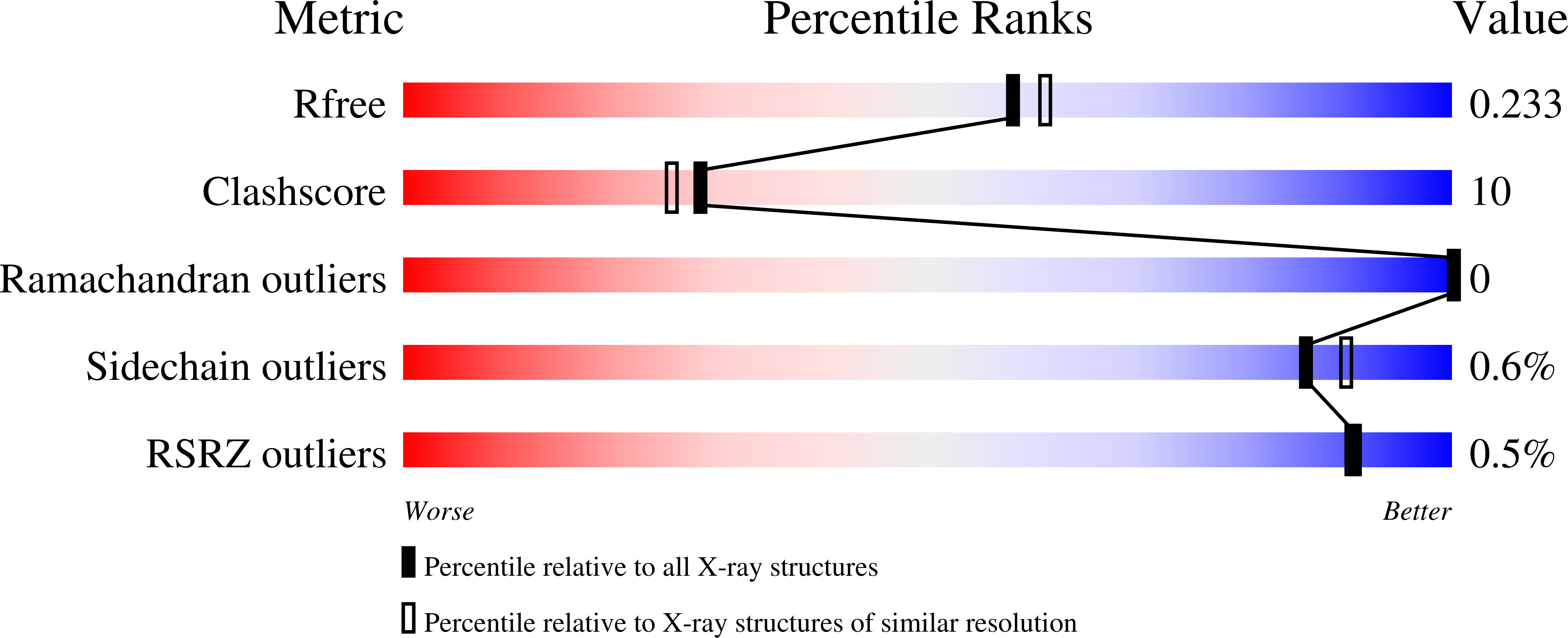
1ULX, 1VGI - PubMed Abstract:
Heme oxygenase (HO) catalyzes physiological heme degradation using O(2) and reducing equivalents to produce biliverdin, iron, and CO. Notably, the HO reaction proceeds without product inhibition by CO, which is generated in the conversion reaction of alpha-hydroxyheme to verdoheme, although CO is known to be a potent inhibitor of HO and other heme proteins. In order to probe how endogenous CO is released from the reaction site, we collected X-ray diffraction data from a crystal of the CO-bound form of the ferrous heme-HO complex in the dark and under illumination by a red laser at approximately 35 K. The difference Fourier map indicates that the CO ligand is partially photodissociated from the heme and that the photolyzed CO is trapped in a hydrophobic cavity adjacent to the heme pocket. This hydrophobic cavity was occupied also by xenon, which is similar to CO in terms of size and properties. Taking account of the affinity of CO for the ferrous verdoheme-HO complex being much weaker than that for the ferrous heme complex, the CO derived from alpha-hydroxyheme would be trapped preferentially in the hydrophobic cavity but not coordinated to the iron of verdoheme. This structural device would ensure the smooth progression of the subsequent reaction, from verdoheme to biliverdin, which requires O(2) binding to verdoheme.
Organizational Affiliation:
Department of Biology, Graduate School of Science, Osaka University, 1-1 Machikaneyama-cho, Toyonaka, Osaka 560-0043, Japan.