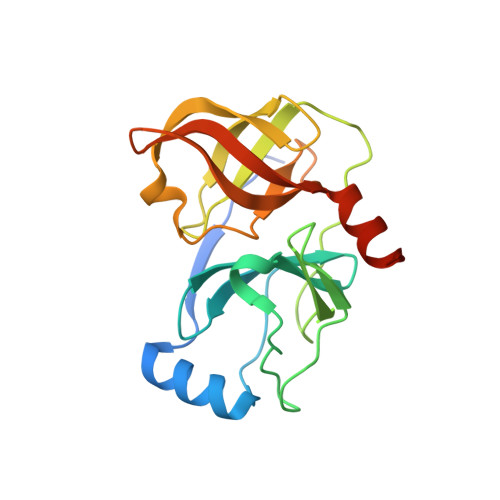
Synthesis and Biological Activity of Macrocyclic Inhibitors of Hepatitis C Virus (HCV) NS3 Protease
Chen, K.X., Njoroge, F.G., Prongay, A., Pichardo, J., Madison, V., Girijavallabhan, V.(2005) Bioorg Med Chem Lett 15: 4475-4478
- PubMed: 16112859
- DOI: https://doi.org/10.1016/j.bmcl.2005.07.033
- Primary Citation of Related Structures:
2A4Q - PubMed Abstract:
The 17-membered phenylalanine-based macrocycle 6 was prepared starting from 3-iodo-phenylalanine. Macrocyclization of alkene phenyl iodide 5 was effected through a palladium-catalyzed Heck reaction. The macrocyclic alpha-ketoamides were active inhibitors of the HCV NS3 protease, with the C-terminal acids and amides being more potent than tert-butyl esters.
Organizational Affiliation:
Schering-Plough Research Institute, 2015 Galloping Hill Road, K-15-3-3545, Kenilworth, NJ 07033, USA. [email protected]