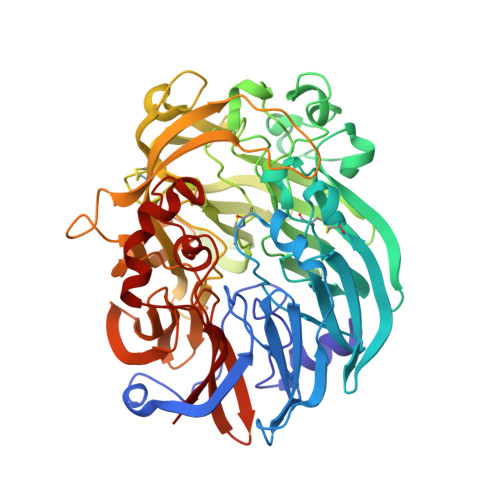
The enzymatic reaction-induced configuration change of the prosthetic group PQQ of methanol dehydrogenase
Li, J., Gan, J.-H., Mathews, F.S., Xia, Z.-X.(2011) Biochem Biophys Res Commun 406: 621-626
- PubMed: 21356200
- DOI: https://doi.org/10.1016/j.bbrc.2011.02.107
- Primary Citation of Related Structures:
2AD6, 2AD7, 2AD8 - PubMed Abstract:
Methanol dehydrogenase is a heterotetrameric enzyme containing the prosthetic group pyrroloquinoline quinone (PQQ), which catalyzes the oxidation of methanol to formaldehyde. The crystal structure of methanol dehydrogenase from Methylophilus W3A1, previously determined at high resolution, exhibits a non-planar configuration of the PQQ ring system and lends support for a hydride transfer mechanism of the enzymatic reaction catalyzed by the enzyme. To investigate why PQQ is in the C5-reduced form and to better understand the catalytic mechanism of the enzyme, three structures of this enzyme in a new crystal form have been determined at higher resolution. Two of the three crystals were grown in the presence of 1 and 50 mM methanol, respectively, both structures of which show non-planar configurations of the PQQ ring system, confirming the previous conclusion; the other was crystallized in the presence of 50 mM ethanol, the structure of which displays a planar ring system for PQQ. Comparison of these structures reveals that the configuration change of PQQ is induced by the enzymatic reaction. The reaction takes place and the C5-reduced PQQ intermediate is produced when the enzyme co-crystallizes with methanol, but the enzymatic reaction does not take place and the PQQ ring retains a planar configuration of the oxidized orthoquinone form when ethanol instead of methanol is present in the crystallization solution.
Organizational Affiliation:
State Key Laboratory of Bio-organic and Natural Products Chemistry, Shanghai Institute of Organic Chemistry, Chinese Academy of Sciences, Shanghai 200032, China.