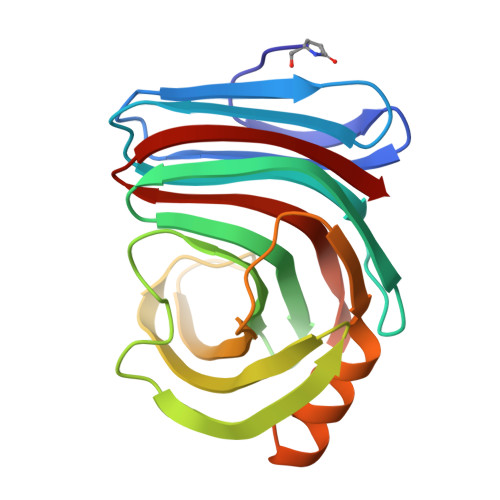
Structure of an orthorhombic form of xylanase II from Trichoderma reesei and analysis of thermal displacement.
Watanabe, N., Akiba, T., Kanai, R., Harata, K.(2006) Acta Crystallogr D Biol Crystallogr 62: 784-792
- PubMed: 16790934
- DOI: https://doi.org/10.1107/S0907444906017379
- Primary Citation of Related Structures:
2DFB, 2DFC - PubMed Abstract:
An orthorhombic crystal of xylanase II from Trichoderma reesei was grown in the presence of sodium iodide. Crystal structures at atomic resolution were determined at 100 and 293 K. Protein molecules were aligned along a crystallographic twofold screw axis, forming a helically extended polymer-like chain mediated by an iodide ion. The iodide ion connected main-chain peptide groups between two adjacent molecules by an N-H...I-...H-N hydrogen-bond bridge, thus contributing to regulation of the molecular arrangement and suppression of the rigid-body motion in the crystal with high diffraction quality. The structure at 293 K showed considerable thermal motion in the loop regions connecting the beta-strands that form the active-site cleft. TLS model analysis of the thermal motion and a comparison between this structure and that at 100 K suggest that the fluctuation of these loop regions is attributable to the hinge-like movement of the beta-strands.
Organizational Affiliation:
Biological Information Research Center, National Institute of Advanced Industrial Science and Technology (AIST), Central 6, 1-1-1 Higashi, Tsukuba, Ibaraki 305-8566, Japan.