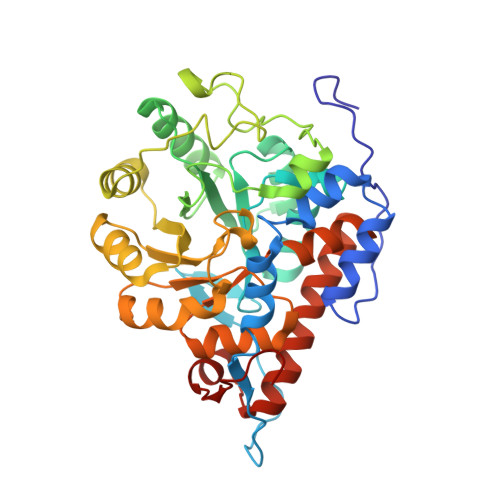
The crystal structure of L-lactate oxidase from Aerococcus viridans at 2.1A resolution reveals the mechanism of strict substrate recognition
Umena, Y., Yorita, K., Matsuoka, T., Kita, A., Fukui, K., Morimoto, Y.(2006) Biochem Biophys Res Commun 350: 249-256
- PubMed: 17007814
- DOI: https://doi.org/10.1016/j.bbrc.2006.09.025
- Primary Citation of Related Structures:
2DU2 - PubMed Abstract:
L-Lactate oxidase (LOX) from Aerococcus viridans is a member of the alpha-hydroxyacid-oxidase flavoenzyme family. We have determined the three-dimensional structure of LOX and revealed the mechanism of substrate recognition. The LOX monomer structure has a typical alpha(8)/beta(8) motif commonly found in other flavin family proteins. A related enzyme, glycolate oxidase, catalyzes the oxidation of glycolate rather than lactate. Comparison of the two enzyme structures highlights the importance of five residues around the FMN prosthetic group of LOX, which act synergistically to discriminate between the l/d configurations of lactate. X-ray crystallography of LOX gave a space group I422 of unit-cell parameters a=b=191.096A, c=194.497A and alpha=beta=gamma=90 degrees with four monomers per asymmetric unit. The four independent monomers display slight structural differences around the active site. Diffraction data were collected, under cryogenic conditions to 2.1A resolution at the synchrotron facilities in Japan.
Organizational Affiliation:
Research Reactor Institute, Kyoto University, Kumatori, Osaka 590-0494, Japan.