The Crystal Structure of Arabidopsis Thaliana Rac7/Rop9: The First Ras Superfamily Gtpase from the Plant Kingdom.
Sormo, C.G., Leiros, I., Brembu, T., Winge, P., Os, V., Bones, A.M.(2006) Phytochemistry 67: 2332
- PubMed: 17005216
- DOI: https://doi.org/10.1016/j.phytochem.2006.08.011
- Primary Citation of Related Structures:
2J0V - PubMed Abstract:
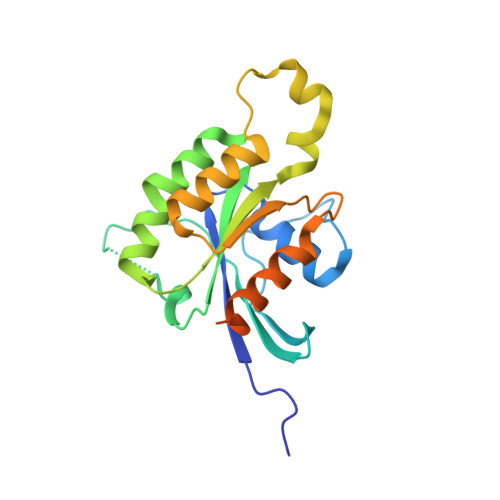
Arabidopsis thaliana RAC/ROP GTPases constitute a plant specific Rho GTPase family in the RAS superfamily, which has been implicated in numerous pivotal signalling cascades in plants. Research has shown that plants in some cases have evolved different modes of regulating Rho GTPase activity as compared to the equivalent systems in animals and yeast. In order to gain structural insight into plant signaling at the molecular level, we have determined the first crystal structure of a RAC-like GTPase belonging to the RAS superfamily from the plant kingdom. The structure of AtRAC7/ROP9 bound to GDP was solved at a resolution of 1.78 A. We have found that the structure of plant Rho GTPases is based upon a conserved G-domain architecture, but structural differences were found concerning the insert region and switch II region of the protein.
Organizational Affiliation:
Department of Biology, Section for Molecular Biology and Biotechnology, Norwegian University of Science and Technology, N-7491 Trondheim, Norway.