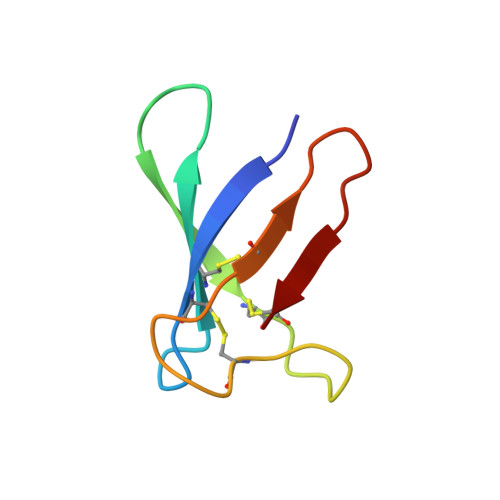
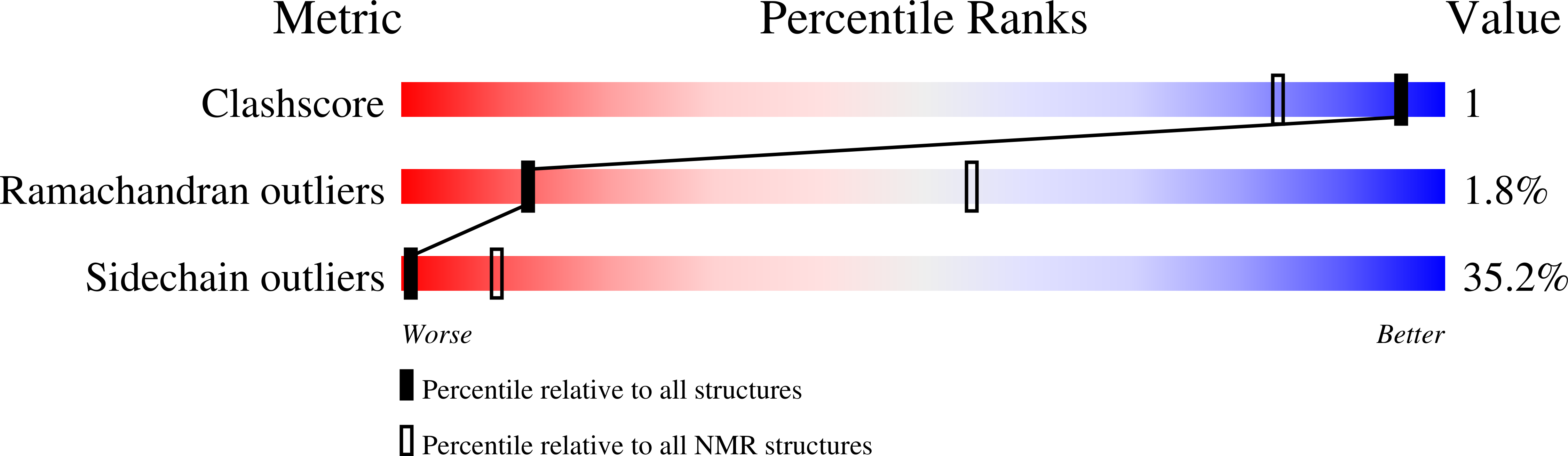"Invisible" Conformers of an Antifungal Disulfide Protein Revealed by Constrained Cold and Heat Unfolding, CEST-NMR Experiments, and Molecular Dynamics Calculations.
Fizil, A., Gaspari, Z., Barna, T., Marx, F., Batta, G.(2015) Chemistry 21: 5136-5144
- PubMed: 25676351
- DOI: https://doi.org/10.1002/chem.201404879
- Primary Citation of Related Structures:
2MHV - PubMed Abstract:
Transition between conformational states in proteins is being recognized as a possible key factor of function. In support of this, hidden dynamic NMR structures were detected in several cases up to populations of a few percent. Here, we show by two- and three-state analysis of thermal unfolding, that the population of hidden states may weight 20-40 % at 298 K in a disulfide-rich protein. In addition, sensitive (15) N-CEST NMR experiments identified a low populated (0.15 %) state that was in slow exchange with the folded PAF protein. Remarkably, other techniques failed to identify the rest of the NMR "dark matter". Comparison of the temperature dependence of chemical shifts from experiments and molecular dynamics calculations suggests that hidden conformers of PAF differ in the loop and terminal regions and are most similar in the evolutionary conserved core. Our observations point to the existence of a complex conformational landscape with multiple conformational states in dynamic equilibrium, with diverse exchange rates presumably responsible for the completely hidden nature of a considerable fraction.
Organizational Affiliation:
Department of Organic Chemistry, Faculty of Science and Technology, University of Debrecen, Egyetem tér 1, 4032 Debrecen (Hungary).