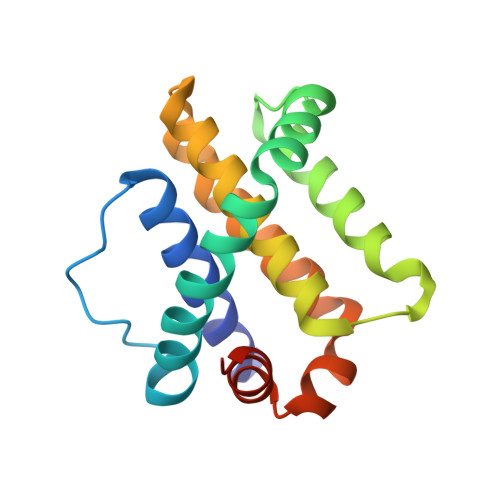
Structural insights into the degradation of Mcl-1 induced by BH3 domains.
Czabotar, P.E., Lee, E.F., van Delft, M.F., Day, C.L., Smith, B.J., Huang, D.C., Fairlie, W.D., Hinds, M.G., Colman, P.M.(2007) Proc Natl Acad Sci U S A 104: 6217-6222
- PubMed: 17389404
- DOI: https://doi.org/10.1073/pnas.0701297104
- Primary Citation of Related Structures:
2JM6, 2NL9, 2NLA - PubMed Abstract:
Apoptosis is held in check by prosurvival proteins of the Bcl-2 family. The distantly related BH3-only proteins bind to and antagonize them, thereby promoting apoptosis. Whereas binding of the BH3-only protein Noxa to prosurvival Mcl-1 induces Mcl-1 degradation by the proteasome, binding of another BH3-only ligand, Bim, elevates Mcl-1 protein levels. We compared the three-dimensional structures of the complexes formed between BH3 peptides of both Bim and Noxa, and we show that a discrete C-terminal sequence of the Noxa BH3 is necessary to instigate Mcl-1 degradation.
Organizational Affiliation:
The Walter and Eliza Hall Institute of Medical Research, Parkville, Victoria 3050, Australia.