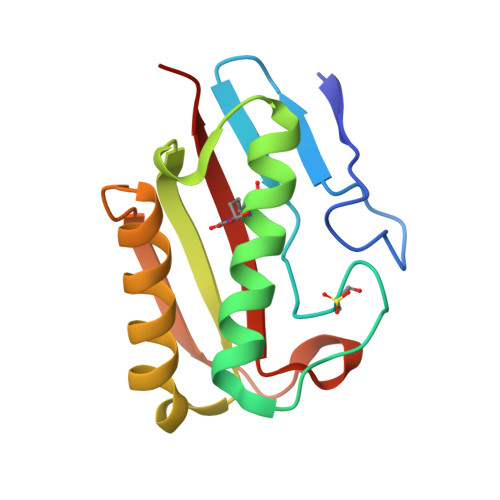
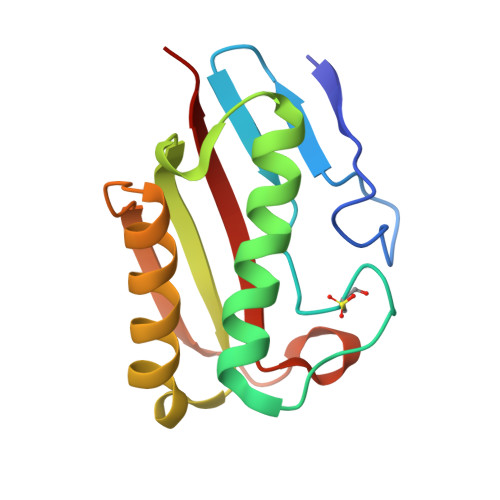
The Crystal Structure of Escherichia Coli Tdcf, a Member of the Highly Conserved Yjgf/Yer057C/Uk114 Family.
Burman, J.D., Stevenson, C.E.M., Sawers, R.G., Lawson, D.M.(2007) BMC Struct Biol 7: 30
- PubMed: 17506874
- DOI: https://doi.org/10.1186/1472-6807-7-30
- Primary Citation of Related Structures:
2UYJ, 2UYK, 2UYN, 2UYP - PubMed Abstract:
The YjgF/YER057c/UK114 family of proteins is widespread in nature, but has as yet no clearly defined biological role. Members of the family exist as homotrimers and are characterised by intersubunit clefts that are delineated by well-conserved residues; these sites are likely to be of functional significance, yet catalytic activity has never been detected for any member of this family. The gene encoding the TdcF protein of E. coli, a YjgF/YER057c/UK114 family member, resides in an operon that strongly suggests a role in the metabolism of 2-ketobutyrate for this protein. We have determined the crystal structure of E. coli TdcF by molecular replacement to a maximum resolution of 1.6 A. Structures are also presented of TdcF complexed with a variety of ligands. The TdcF structure closely resembles those of all YjgF/YER057c/UK114 family members determined thus far. It has the trimeric quaternary structure and intersubunit cavities characteristic of this family of proteins. We show that TdcF is capable of binding several low molecular weight metabolites bearing a carboxylate group, although the interaction with 2-ketobutyrate appears to be the most well defined. These observations may be indicative of a role for TdcF in sensing this potentially toxic metabolite.
Organizational Affiliation:
Department of Biological Chemistry, John Innes Centre, Norwich, UK. [email protected]