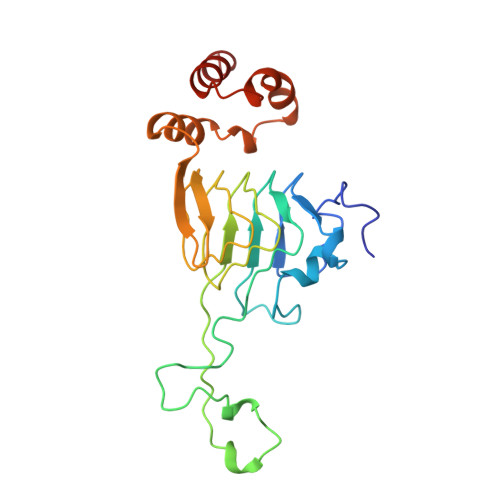
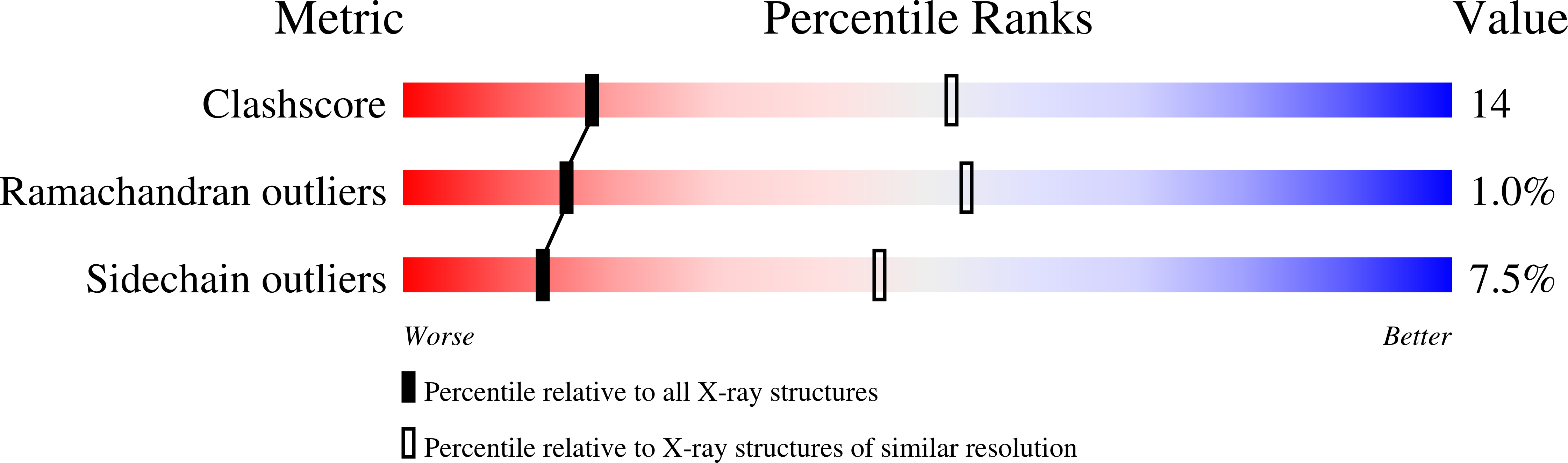Structure of the hexapeptide xenobiotic acetyltransferase from Pseudomonas aeruginosa.
Beaman, T.W., Sugantino, M., Roderick, S.L.(1998) Biochemistry 37: 6689-6696
- PubMed: 9578552
- DOI: https://doi.org/10.1021/bi980106v
- Primary Citation of Related Structures:
1XAT, 2XAT - PubMed Abstract:
The crystal structure of the xenobiotic acetyltransferase from Pseudomonas aeruginosa PA103 (PaXAT) has been determined, as well as that of its complex with the substrate chloramphenicol and the cofactor analogue desulfo-coenzyme A. PaXAT is a member of the large hexapeptide acyltransferase family of enzymes that display tandem repeated copies of a six-residue hexapeptide repeat sequence motif encoding a left-handed parallel beta helix (L betaH) structural domain. The xenobiotic acetyltransferase class of hexapeptide acyltransferases is composed of microbial enzymes that utilize acetyl-CoA to acylate a variety of hydroxyl-bearing acceptors. The active site of trimeric PaXAT is a short tunnel into which chloramphenicol and the cofactor analogue desulfo-CoA project from opposite ends. This tunnel is formed by the flat parallel beta sheets of two separate L betaH domains and an extended 39-residue loop. His 79 of the extended loop forms hydrogen bonds from its imidazole NE2 atom to the 3-hydroxyl group of chloramphenicol and from its ND1 group to the peptide oxygen of Thr 86. The interactions of this histidine residue are similar to those found in the structurally unrelated type III chloramphenicol acetyltransferase and suggest that His 79 of PaXAT may be similarly positioned and tautomerically stabilized to serve as a general base catalyst.
Organizational Affiliation:
Department of Biochemistry, Albert Einstein College of Medicine, Bronx, New York 10461, USA.