Fam3B Pander and Fam3C Ilei Represent a Distinct Class of Signaling Molecules with a Non-Cytokine-Like Fold.
Johansson, P., Bernstrom, J., Gorman, T., Oster, L., Backstrom, S., Schweikart, F., Xu, B., Xue, Y., Schiavone, L.H.(2013) Structure 21: 306
- PubMed: 23333428
- DOI: https://doi.org/10.1016/j.str.2012.12.009
- Primary Citation of Related Structures:
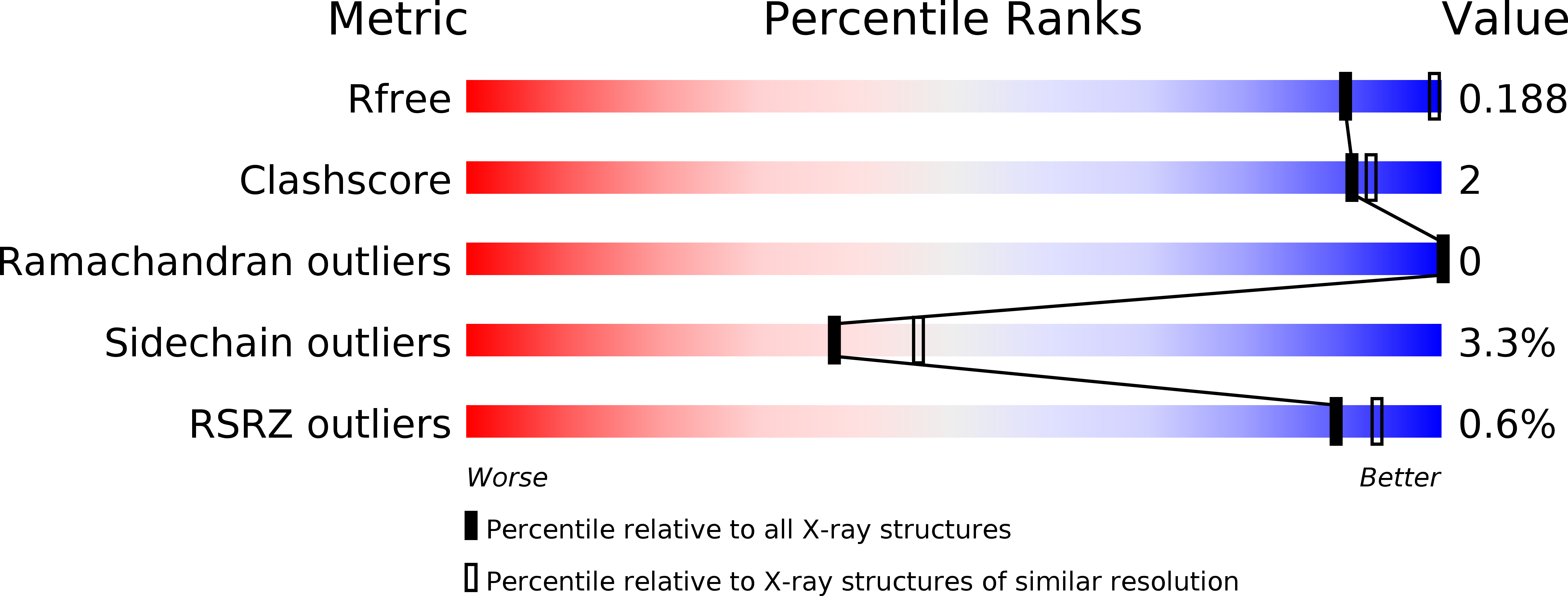
2YOP, 2YOQ - PubMed Abstract:
The FAM3 superfamily is predicted to contain classical four-helix bundle cytokines, featuring a typical up-up-down-down fold. Two members of FAM3 have been extensively studied. FAM3B PANDER has been shown to regulate glucose homeostasis and β cell function, whereas the homologous FAM3C ILEI has been shown to be involved in epithelial-mesenchymal transition and cancer. Here, we present a three-dimensional structure of a FAM3 protein, murine PANDER. Contrary to previous suggestions, PANDER exhibits a globular β-β-α fold. The structure is composed of two antiparallel β sheets lined by three short helices packing to form a highly conserved water-filled cavity. The fold shares no relation to the predicted four-helix cytokines but is conserved throughout the FAM3 superfamily. The available biological data and the unexpected new fold indicate that FAM3 PANDER and ILEI could represent a new structural class of signaling molecules, with a different mode of action compared to the traditional four-helix bundle cytokines.
Organizational Affiliation:
Structure and Biophysics, Discovery Sciences, AstraZeneca, Mölndal 431-83, Sweden.