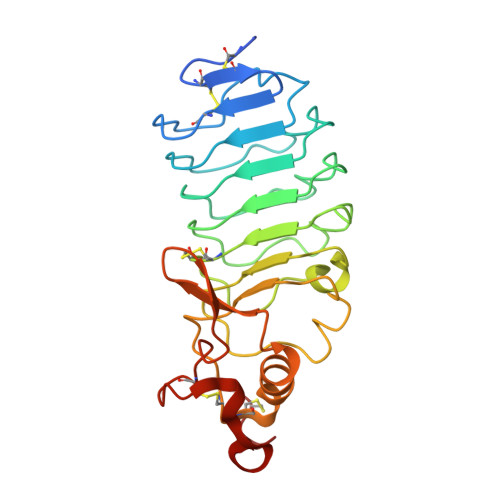
Antigen recognition by variable lymphocyte receptors.
Han, B.W., Herrin, B.R., Cooper, M.D., Wilson, I.A.(2008) Science 321: 1834-1837
- PubMed: 18818359
- DOI: https://doi.org/10.1126/science.1162484
- Primary Citation of Related Structures:
3E6J - PubMed Abstract:
Variable lymphocyte receptors (VLRs) rather than antibodies play the primary role in recognition of antigens in the adaptive immune system of jawless vertebrates. Combinatorial assembly of leucine-rich repeat (LRR) gene segments achieves the required repertoire for antigen recognition. We have determined a crystal structure for a VLR-antigen complex, VLR RBC36 in complex with the H-antigen trisaccharide from human blood type O erythrocytes, at 1.67 angstrom resolution. RBC36 binds the H-trisaccharide on the concave surface of the LRR modules of the solenoid structure where three key hydrophilic residues, multiple van der Waals interactions, and the highly variable insert of the carboxyl-terminal LRR module determine antigen recognition and specificity. The concave surface assembled from the most highly variable regions of the LRRs, along with diversity in the sequence and length of the highly variable insert, can account for the recognition of diverse antigens by VLRs.
Organizational Affiliation:
Department of Molecular Biology, Scripps Research Institute, La Jolla, CA 92037, USA.