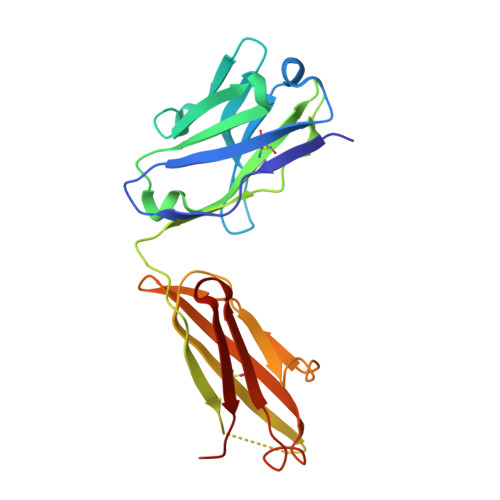
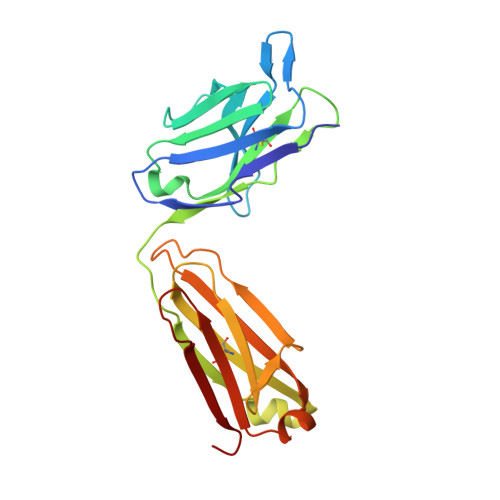
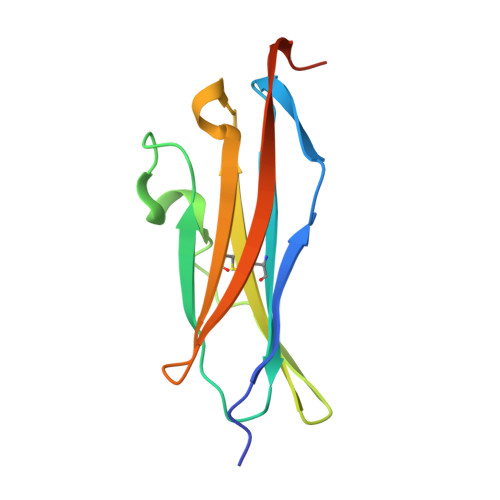
The Structural Basis for the Function of Two Anti-VEGF Receptor 2 Antibodies.
Franklin, M.C., Navarro, E.C., Wang, Y., Patel, S., Singh, P., Zhang, Y., Persaud, K., Bari, A., Griffith, H., Shen, L., Balderes, P., Kussie, P.(2011) Structure 19: 1097-1107
- PubMed: 21827946
- DOI: https://doi.org/10.1016/j.str.2011.01.019
- Primary Citation of Related Structures:
3S34, 3S35, 3S36, 3S37 - PubMed Abstract:
The anti-VEGF receptor 2 antibody IMC-1121B is a promising antiangiogenic drug being tested for treatment of breast and gastric cancer. We have determined the structure of the 1121B Fab fragment in complex with domain 3 of VEGFR2, as well as the structure of a different neutralizing anti-VEGFR2 antibody, 6.64, also in complex with VEGFR2 domain 3. The two Fab fragments bind at opposite ends of VEGFR2 domain 3; 1121B directly blocks VEGF binding, whereas 6.64 may prevent receptor dimerization by perturbing the domain 3:domain 4 interface. Mutagenesis reveals that residues essential for VEGF, 1121B, and 6.64 binding are nonoverlapping among the three contact patches.
Organizational Affiliation:
Department of Immunology, ImClone Systems, New York, NY 10014, USA. [email protected]