Crystal structure of human CYP7A1 in complex with 7-ketocholesterol
Strushkevich, N., Tempel, W., MacKenzie, F., Wernimont, A.K., Arrowsmith, C.H., Edwards, A.M., Bountra, C., Weigelt, J., Usanov, S.A., Park, H.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Cholesterol 7-alpha-monooxygenase | 491 | Homo sapiens | Mutation(s): 1 Gene Names: CYP7A1, CYP7 EC: 1.14.13.17 (PDB Primary Data), 1.14.14.26 (UniProt), 1.14.14.23 (UniProt) | 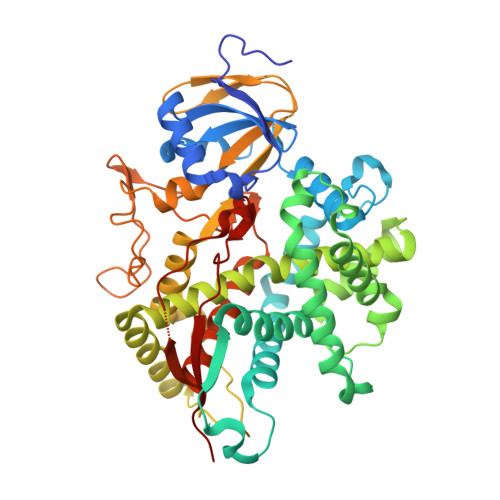 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P22680 (Homo sapiens) Explore P22680 Go to UniProtKB: P22680 | |||||
PHAROS: P22680 GTEx: ENSG00000167910 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P22680 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 4 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| HEM Query on HEM | C [auth A], M [auth B] | PROTOPORPHYRIN IX CONTAINING FE C34 H32 Fe N4 O4 KABFMIBPWCXCRK-RGGAHWMASA-L |  | ||
| 0GV Query on 0GV | D [auth A], N [auth B] | (3beta,8alpha,9beta)-3-hydroxycholest-5-en-7-one C27 H44 O2 YIKKMWSQVKJCOP-ABXCMAEBSA-N |  | ||
| SO4 Query on SO4 | E [auth A] F [auth A] G [auth A] O [auth B] P [auth B] | SULFATE ION O4 S QAOWNCQODCNURD-UHFFFAOYSA-L |  | ||
| UNX Query on UNX | H [auth A] I [auth A] J [auth A] K [auth A] L [auth A] | UNKNOWN ATOM OR ION X |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 55.618 | α = 66.33 |
| b = 74.22 | β = 75.53 |
| c = 87.933 | γ = 69.62 |
| Software Name | Purpose |
|---|---|
| DENZO | data reduction |
| SCALEPACK | data scaling |
| PHASER | phasing |
| REFMAC | refinement |
| PDB_EXTRACT | data extraction |
| HKL-2000 | data reduction |
| HKL-2000 | data scaling |