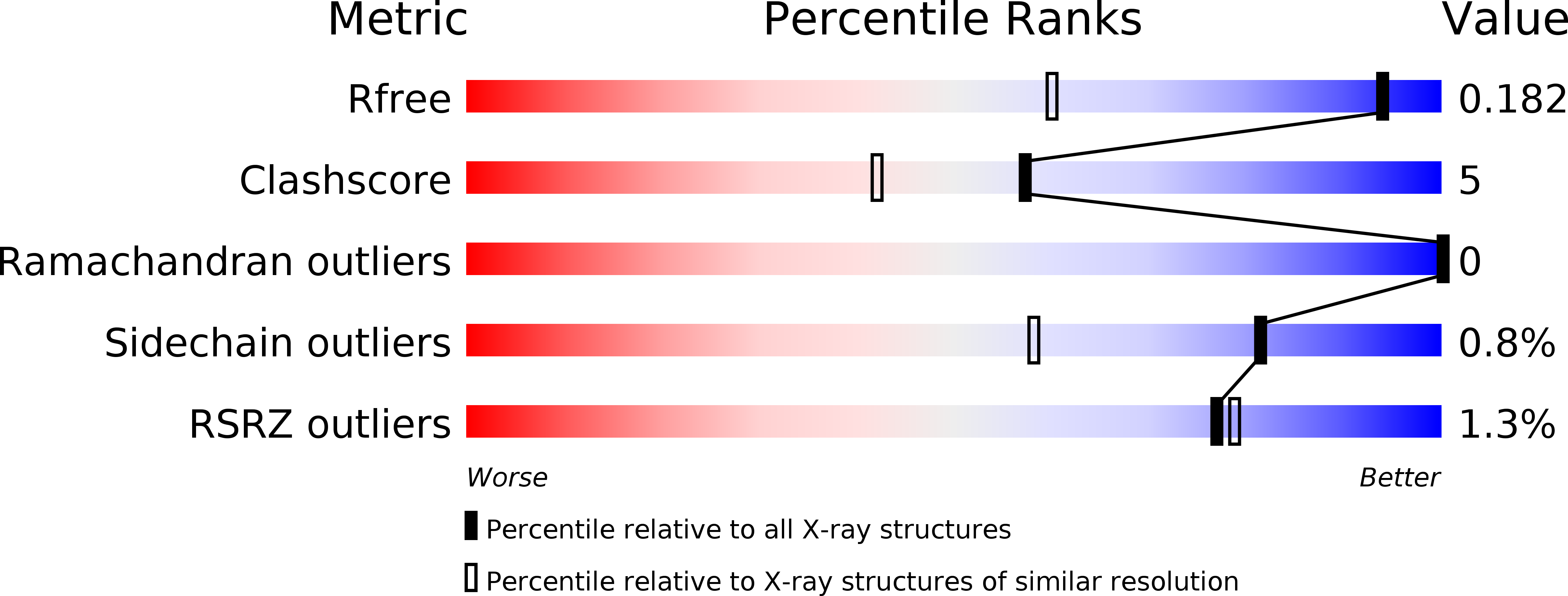
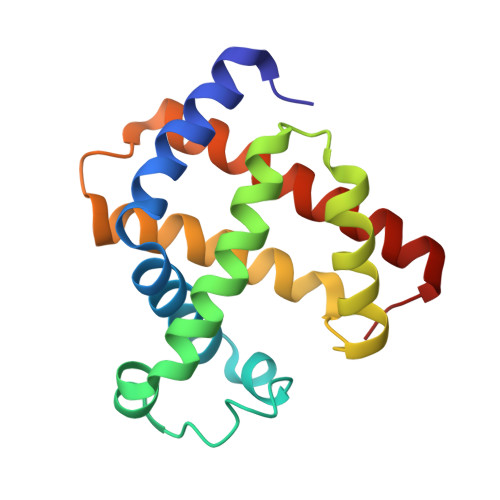
Co(II)/Co(I) reduction-induced axial histidine-flipping in myoglobin reconstituted with a cobalt tetradehydrocorrin as a methionine synthase model.
Hayashi, T., Morita, Y., Mizohata, E., Oohora, K., Ohbayashi, J., Inoue, T., Hisaeda, Y.(2014) Chem Commun (Camb) 50: 12560-12563
- PubMed: 25197974
- DOI: https://doi.org/10.1039/c4cc05448b
- Primary Citation of Related Structures:
3WFT, 3WFU - PubMed Abstract:
A conjugate between apomyoglobin and cobalt tetradehydrocorrin was prepared to replicate the coordination behavior of cob(I)alamin in methionine synthase. X-ray crystallography reveals that the tetra-coordinated Co(I) species is formed through the cleavage of the axial Co-His93 ligation after the reduction of the penta-coordinated Co(II) cofactor in the heme pocket.
Organizational Affiliation:
Department of Applied Chemistry, Graduate School of Engineering, Osaka University, Suita 565-0871, Japan. [email protected].