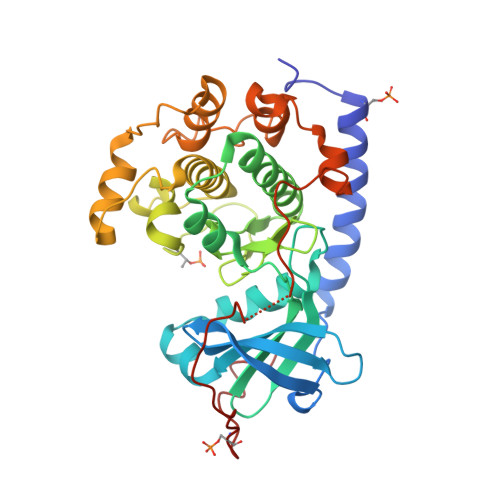
Mutants of protein kinase A that mimic the ATP-binding site of Aurora kinase
Pflug, A., de Oliveira, T.M., Bossemeyer, D., Engh, R.A.(2011) Biochem J 440: 85-93
- PubMed: 21774789
- DOI: https://doi.org/10.1042/BJ20110592
- Primary Citation of Related Structures:
3AMA, 3AMB - PubMed Abstract:
We describe in the present paper mutations of the catalytic subunit α of PKA (protein kinase A) that introduce amino acid side chains into the ATP-binding site and progressively transform the pocket to mimic that of Aurora protein kinases. The resultant PKA variants are enzymatically active and exhibit high affinity for ATP site inhibitors that are specific for Aurora kinases. These features make the Aurora-chimaeric PKA a valuable tool for structure-based drug discovery tasks. Analysis of crystal structures of the chimaera reveal the roles for individual amino acid residues in the binding of a variety of inhibitors, offering key insights into selectivity mechanisms. Furthermore, the high affinity for Aurora kinase-specific inhibitors, combined with the favourable crystallizability properties of PKA, allow rapid determination of inhibitor complex structures at an atomic resolution. We demonstrate the utility of the Aurora-chimaeric PKA by measuring binding kinetics for three Aurora kinase-specific inhibitors, and present the X-ray structures of the chimaeric enzyme in complex with VX-680 (MK-0457) and JNJ-7706621 [Aurora kinase/CDK (cyclin-dependent kinase) inhibitor].
Organizational Affiliation:
Norwegian Structural Biology Centre, Department of Chemistry, University of Tromsø, N-9037 Tromsø, Norway.