Combined Inhibitor Free-Energy Landscape and Structural Analysis Reports on the Mannosidase Conformational Coordinate.
Williams, R.J., Iglesias-Fernandez, J., Stepper, J., Jackson, A., Thompson, A.J., Lowe, E.C., White, J.M., Gilbert, H.J., Rovira, C., Davies, G.J., Williams, S.J.(2014) Angew Chem Int Ed Engl 53: 1087
- PubMed: 24339341
- DOI: https://doi.org/10.1002/anie.201308334
- Primary Citation of Related Structures:
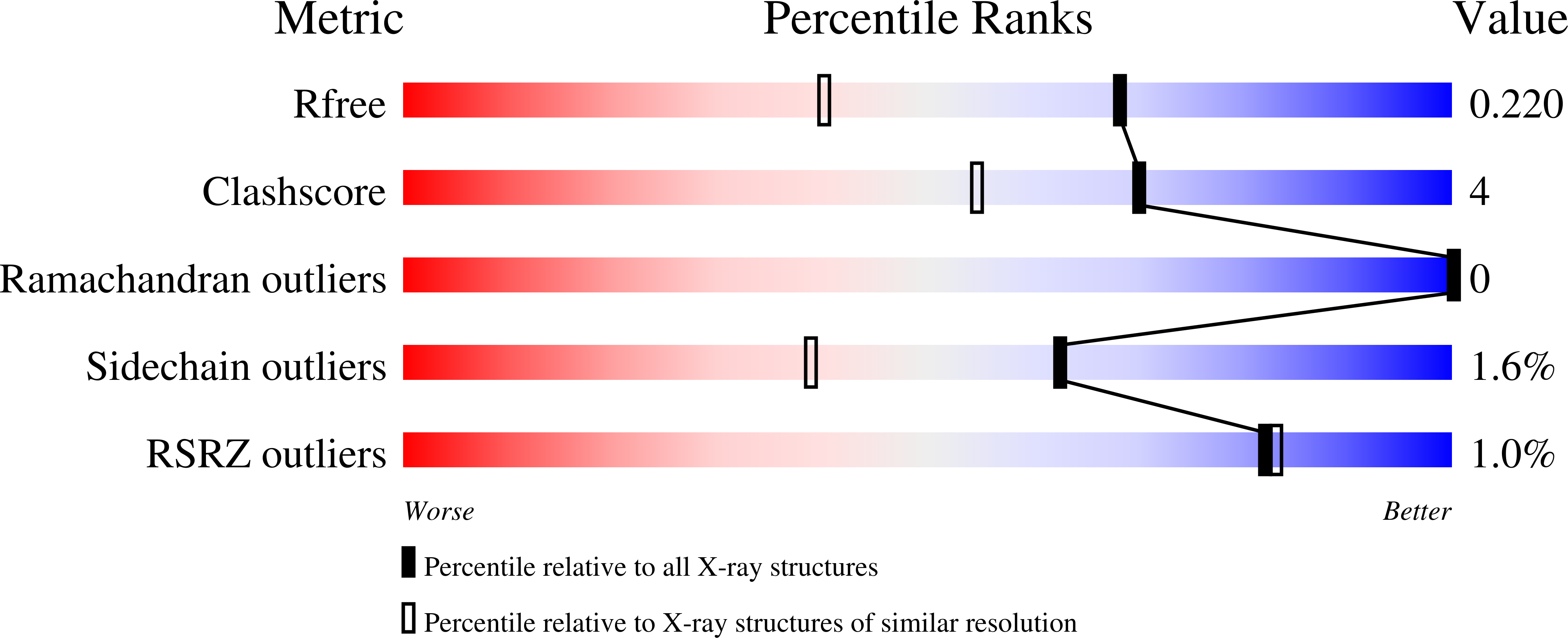
4CD4, 4CD5, 4CD6, 4CD7, 4CD8 - PubMed Abstract:
Mannosidases catalyze the hydrolysis of a diverse range of polysaccharides and glycoconjugates, and the various sequence-based mannosidase families have evolved ingenious strategies to overcome the stereoelectronic challenges of mannoside chemistry. Using a combination of computational chemistry, inhibitor design and synthesis, and X-ray crystallography of inhibitor/enzyme complexes, it is demonstrated that mannoimidazole-type inhibitors are energetically poised to report faithfully on mannosidase transition-state conformation, and provide direct evidence for the conformational itinerary used by diverse mannosidases, including β-mannanases from families GH26 and GH113. Isofagomine-type inhibitors are poor mimics of transition-state conformation, owing to the high energy barriers that must be crossed to attain mechanistically relevant conformations, however, these sugar-shaped heterocycles allow the acquisition of ternary complexes that span the active site, thus providing valuable insight into active-site residues involved in substrate recognition.
Organizational Affiliation:
School of Chemistry and Bio21 Molecular Science and Biotechnology Institute, University of Melbourne, Parkville, Vic 3010 (Australia).