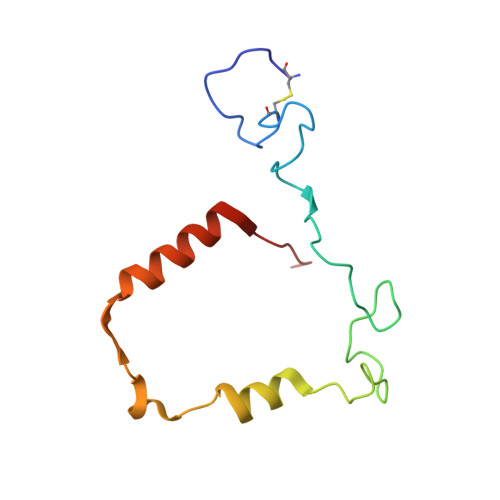
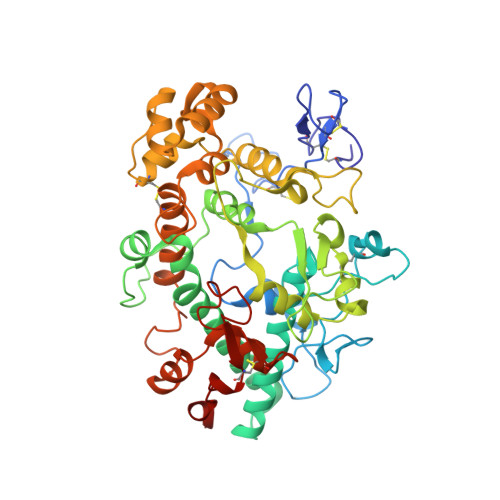
Deconstruction of activity-dependent covalent modification of heme in human neutrophil myeloperoxidase by multistage mass spectrometry (MS(4)).
Geoghegan, K.F., Varghese, A.H., Feng, X., Bessire, A.J., Conboy, J.J., Ruggeri, R.B., Ahn, K., Spath, S.N., Filippov, S.V., Conrad, S.J., Carpino, P.A., Guimaraes, C.R., Vajdos, F.F.(2012) Biochemistry 51: 2065-2077
- PubMed: 22352991
- DOI: https://doi.org/10.1021/bi201872j
- Primary Citation of Related Structures:
4DL1 - PubMed Abstract:
Myeloperoxidase (MPO) is known to be inactivated and covalently modified by treatment with hydrogen peroxide and agents similar to 3-(2-ethoxypropyl)-2-thioxo-2,3-dihydro-1H-purin-6(9H)-one (1), a 254.08 Da derivative of 2-thioxanthine. Peptide mapping by liquid chromatography and mass spectrometry detected modification by 1 in a labile peptide-heme-peptide fragment of the enzyme, accompanied by a mass increase of 252.08 Da. The loss of two hydrogen atoms was consistent with mechanism-based oxidative coupling. Multistage mass spectrometry (MS(4)) of the modified fragment in an ion trap/Orbitrap spectrometer demonstrated that 1 was coupled directly to heme. Use of a 10 amu window delivered the full isotopic envelope of each precursor ion to collision-induced dissociation, preserving definitive isotopic profiles for iron-containing fragments through successive steps of multistage mass spectrometry. Iron isotope signatures and accurate mass measurements supported the structural assignments. Crystallographic analysis confirmed linkage between the methyl substituent of the heme pyrrole D ring and the sulfur atom of 1. The final orientation of 1 perpendicular to the plane of the heme ring suggested a mechanism consisting of two consecutive one-electron oxidations of 1 by MPO. Multistage mass spectrometry using stage-specific collision energies permits stepwise deconstruction of modifications of heme enzymes containing covalent links between the heme group and the polypeptide chain.
Organizational Affiliation:
Pfizer Worldwide Research, Groton, Connecticut 06340, United States. [email protected]