The Glucose Acceptor site of lacZ beta-galactosidase for the synthesis of allolactose - the natural inducer of the lac operon
Wheatley, R.W., Lo, S., Janzcewicz, L.J., Dugdale, M.L., Huber, R.E.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Biological assembly 1 assigned by authors and generated by PISA (software)
Macromolecule Content
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Beta-galactosidase | 1,052 | Escherichia coli K-12 | Mutation(s): 1 Gene Names: b0344, JW0335, lacZ EC: 3.2.1.23 | 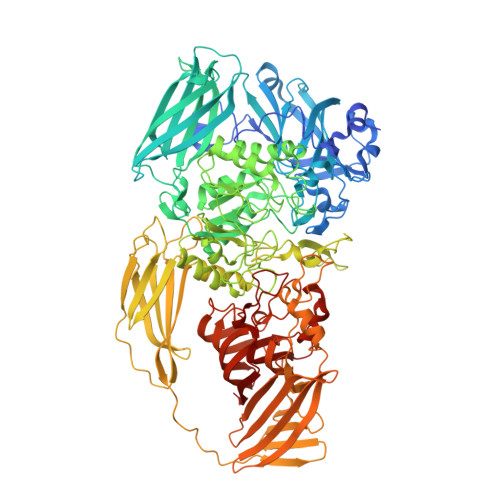 | |
UniProt | |||||
Find proteins for P00722 (Escherichia coli (strain K12)) Explore P00722 Go to UniProtKB: P00722 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P00722 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 150.57 | α = 90 |
| b = 167.731 | β = 90 |
| c = 201.64 | γ = 90 |
| Software Name | Purpose |
|---|---|
| REFMAC | refinement |
| CNS | refinement |
| ADSC | data collection |
| MOSFLM | data reduction |
| SCALA | data scaling |
| CNS | phasing |
RCSB PDB Core Operations are funded by the U.S. National Science Foundation (DBI-2321666), the US Department of Energy (DE-SC0019749), and the National Cancer Institute, National Institute of Allergy and Infectious Diseases, and National Institute of General Medical Sciences of the National Institutes of Health under grant R01GM157729.





