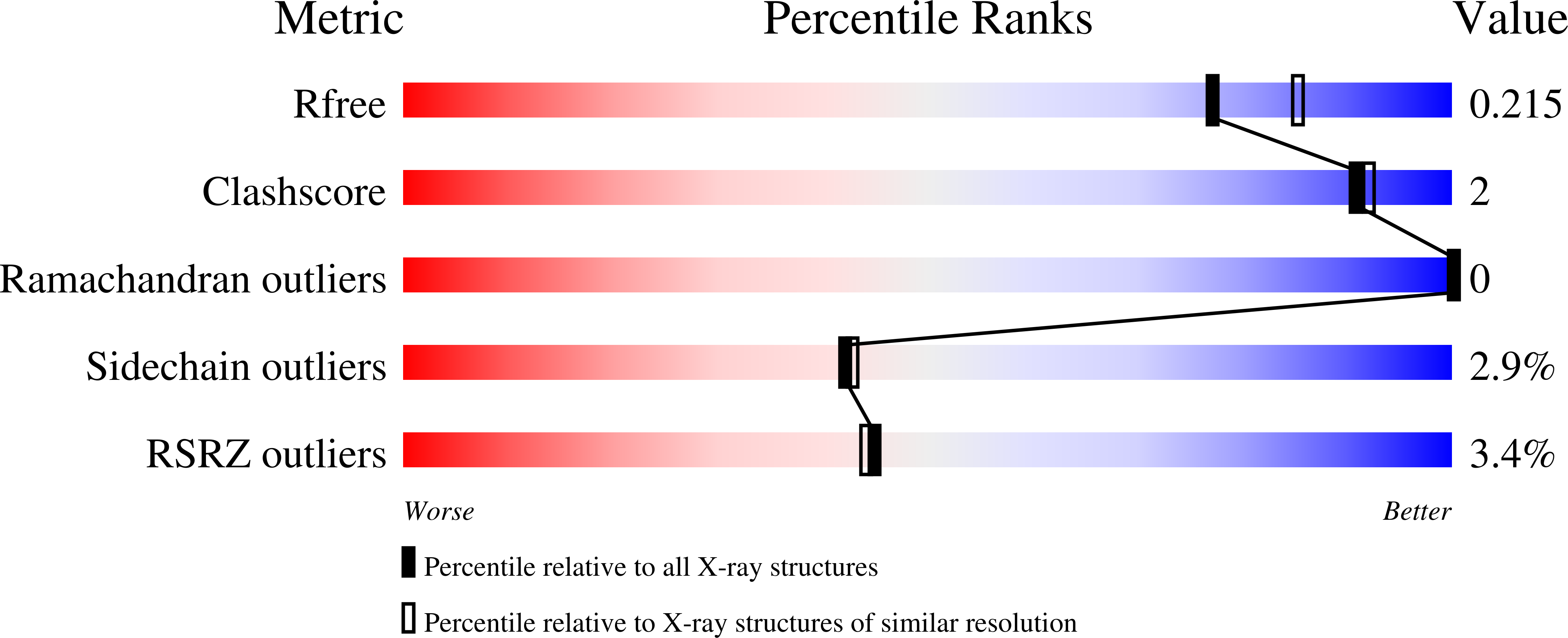
Kainate induces various domain closures in AMPA and kainate receptors.
Venskutonyte, R., Frydenvang, K., Hald, H., Rabassa, A.C., Gajhede, M., Ahring, P.K., Kastrup, J.S.(2012) Neurochem Int 61: 536-545
- PubMed: 22425692
- DOI: https://doi.org/10.1016/j.neuint.2012.02.016
- Primary Citation of Related Structures:
4E0W, 4E0X - PubMed Abstract:
Ionotropic glutamate receptors are key players in fast excitatory synaptic transmission within the central nervous system. These receptors have been divided into three subfamilies: the N-methyl-d-aspartic acid (NMDA), 2-amino-3-(3-hydroxy-5-methyl-4-isoxazolyl)propionic acid (AMPA) and kainate receptors. Kainate has previously been crystallized with the ligand binding domain (LBD) of AMPA receptors (GluA2 and GluA4) and kainate receptors (GluK1 and GluK2). Here, we report the structures of the kainate receptor GluK3 LBD in complex with kainate and GluK1 LBD in complex with kainate in the absence of glycerol. Kainate introduces a conformational change in GluK3 LBD comparable to that of GluK2, but different from the conformational changes induced in GluA2 and GluK1. Compared to their domain closures in a glutamate bound state, GluA2 and GluK1 become more open and kainate induces a domain closure of 60% and 62%, respectively, relative to glutamate (100%). In GluK2 and GluK3 with kainate, the domain closure is 88% and 83%, respectively. In previously determined structures of GluK1 LBD in complex with kainate, glycerol is present in the binding site where it bridges interlobe residues and thus, might contribute to the large domain opening. However, the structure of GluK1 LBD with kainate in the absence of glycerol confirms that the observed domain closure is not an artifact of crystallization conditions. Comparison of the LBD structures with glutamate and kainate reveals that contacts are lost upon binding of kainate in the three kainate receptors, which is in contrast to the AMPA receptors where similar contacts are seen. It was revealed by patch clamp electrophysiology studies that kainate is a partial agonist at GluK1 with 36% efficacy compared to glutamate, which is in between the published efficacies of kainate at GluK2 and AMPA receptors. The ranking of efficacies seems to correlate with LBD domain closures.
Organizational Affiliation:
Department of Drug Design and Pharmacology, Faculty of Health and Medical Sciences, University of Copenhagen, Universitetsparken 2, DK-2100 Copenhagen, Denmark.