Structural basis of sulfonation of the glycopeptide antibiotic scaffold by the sulfotransferase StaL
Shi, R., Munger, C., Kalan, L., Sulea, T., Wright, G.D., Cygler, M.(2012) Proc Natl Acad Sci U S A
Experimental Data Snapshot
Starting Model: experimental
View more details
(2012) Proc Natl Acad Sci U S A
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| StaL | 286 | Streptomyces toyocaensis | Mutation(s): 0 Gene Names: stal EC: 2.8.2.36 | 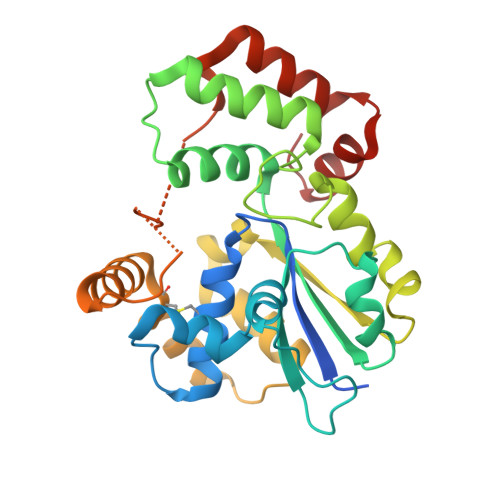 | |
UniProt | |||||
Find proteins for Q8KLM3 (Streptomyces toyocaensis) Explore Q8KLM3 Go to UniProtKB: Q8KLM3 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q8KLM3 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar proteins by: Sequence | 3D Structure
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| desulfo-A47934 | 7 | Streptomyces toyocaensis | Mutation(s): 0 | 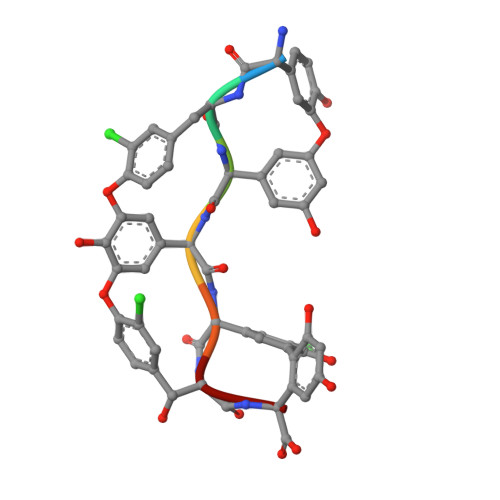 | |
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| A3P Query on A3P | D [auth A] | ADENOSINE-3'-5'-DIPHOSPHATE C10 H15 N5 O10 P2 WHTCPDAXWFLDIH-KQYNXXCUSA-N |  | ||
| Modified Residues 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Type | Formula | 2D Diagram | Parent |
| 3MY Query on 3MY | C | D-PEPTIDE LINKING | C9 H10 Cl N O3 |  | TYR |
| OMY Query on OMY | C | L-PEPTIDE LINKING | C9 H10 Cl N O4 |  | TYR |
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| ID | Chains | Name | Type/Class | 2D Diagram | 3D Interactions |
| PRD_000812 Query on PRD_000812 | C | desulfo-A47934 | Glycopeptide / Antibiotic |  | |
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 53.442 | α = 90 |
| b = 82.583 | β = 90 |
| c = 123.076 | γ = 90 |
| Software Name | Purpose |
|---|---|
| DENZO | data reduction |
| SCALEPACK | data scaling |
| REFMAC | refinement |
| PDB_EXTRACT | data extraction |
| HKL-2000 | data reduction |
| HKL-2000 | data scaling |
| MOLREP | phasing |