Structural principles for Alpha-neurotoxin binding to and selectivity among nicotinic receptors
Li, S.X., Cheng, K., Gomoto, R., Bren, N., Huang, S., Sine, S., Chen, L.To be published.
Experimental Data Snapshot
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Alpha7 nicotinic receptor chimera | 202 | Homo sapiens, Lymnaea stagnalis This entity is chimeric | Mutation(s): 0 | 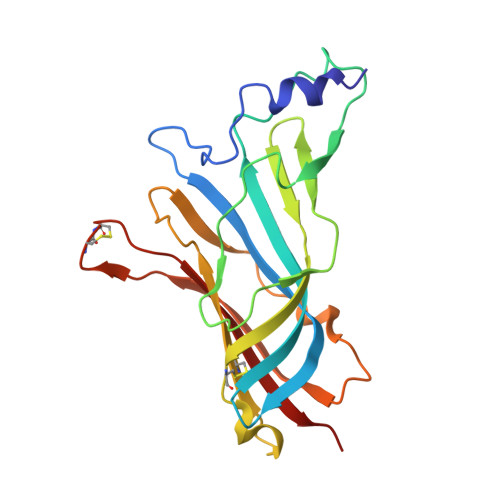 | |
UniProt | |||||
Find proteins for P58154 (Lymnaea stagnalis) Explore P58154 Go to UniProtKB: P58154 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P58154 | ||||
Glycosylation | |||||
| Glycosylation Sites: 2 | |||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Alpha-bungarotoxin isoform V31 | 73 | Bungarus multicinctus | Mutation(s): 0 | 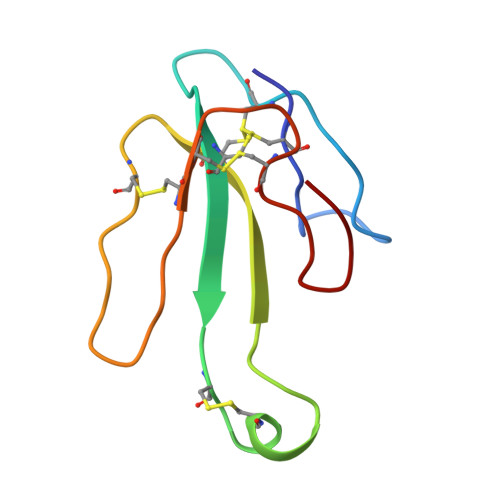 | |
UniProt | |||||
Find proteins for P60616 (Bungarus multicinctus) Explore P60616 Go to UniProtKB: P60616 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P60616 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| NAG Query on NAG | L [auth A] M [auth A] N [auth B] O [auth C] P [auth C] | 2-acetamido-2-deoxy-beta-D-glucopyranose C8 H15 N O6 OVRNDRQMDRJTHS-FMDGEEDCSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 142.15 | α = 90 |
| b = 142.15 | β = 90 |
| c = 518.135 | γ = 120 |
| Software Name | Purpose |
|---|---|
| CNS | refinement |