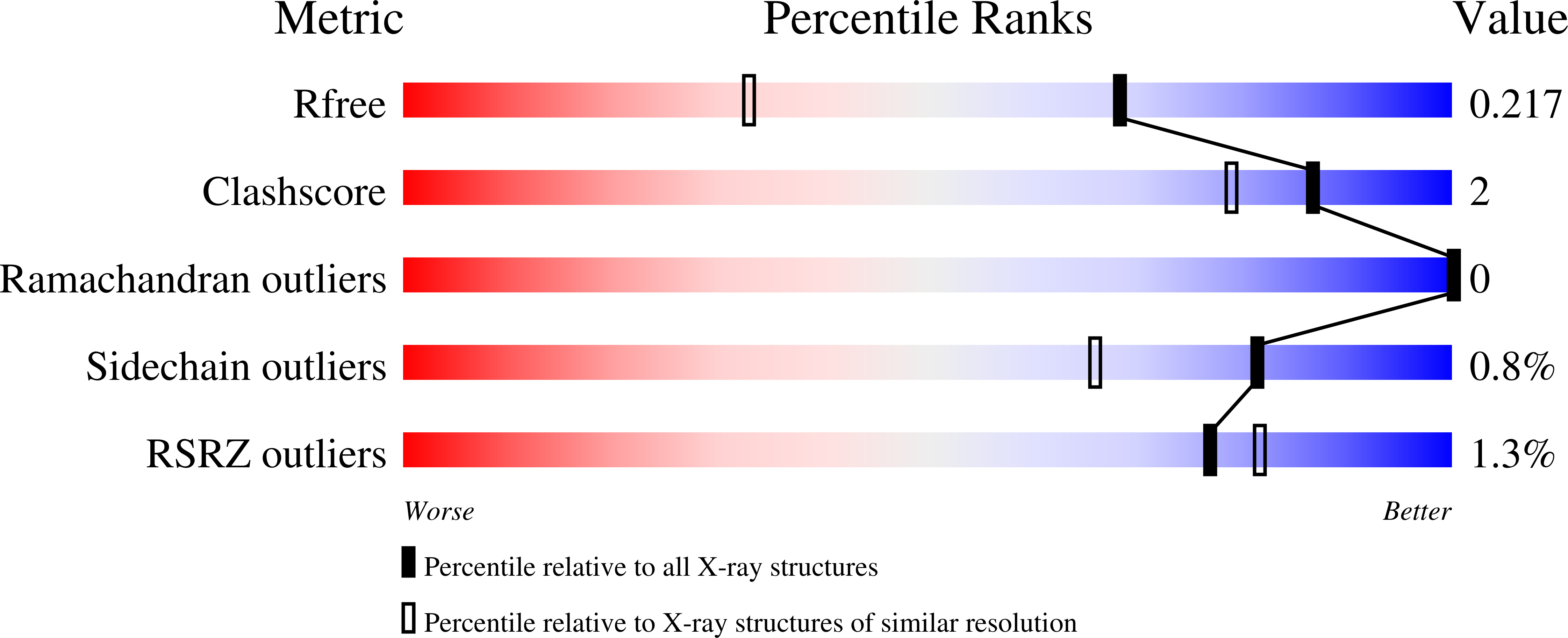
Reversal of the substrate specificity of CMP N-glycosidase to dCMP.
Sikowitz, M.D., Cooper, L.E., Begley, T.P., Kaminski, P.A., Ealick, S.E.(2013) Biochemistry 52: 4037-4047
- PubMed: 23659472
- DOI: https://doi.org/10.1021/bi400316p
- Primary Citation of Related Structures:
4JEL, 4JEM, 4JEN - PubMed Abstract:
MilB is a CMP hydrolase involved in the early steps of biosynthesis of the antifungal compound mildiomycin. An enzyme from the bacimethrin biosynthetic pathway, BcmB, is closely related to MilB in both sequence and function. These two enzymes belong to the nucleoside 2'-deoxyribosyltransferase (NDT) superfamily. NDTs catalyze N-glycosidic bond cleavage of 2'-deoxynucleosides via a covalent 2-deoxyribosyl-enzyme intermediate. Conservation of key active site residues suggests that members of the NDT superfamily share a common mechanism; however, the enzymes differ in their substrate preferences. Substrates vary in the type of nucleobase, the presence or absence of a 2'-hydroxyl group, and the presence or absence of a 5'-phosphate group. We have determined the structures of MilB and BcmB and compared them to previously determined structures of NDT superfamily members. The comparisons reveal how these enzymes differentiate between ribosyl and deoxyribosyl nucleotides or nucleosides and among different nucleobases. The 1.6 Å structure of the MilB-CMP complex reveals an active site feature that is not obvious from comparisons of sequence alone. MilB and BcmB that prefer substrates containing 2'-ribosyl groups have a phenylalanine positioned in the active site, whereas NDT family members with a preference for 2'-deoxyribosyl groups have a tyrosine residue. Further studies show that the phenylalanine is critical for the specificity of MilB and BcmB toward CMP, and mutation of this phenylalanine residue to tyrosine results in a 1000-fold reversal of substrate specificity from CMP to dCMP.
Organizational Affiliation:
Department of Chemistry and Chemical Biology, Cornell University, Ithaca, NY 14853, USA.