Rational HIV immunogen design to target specific germline B cell receptors.
Jardine, J., Julien, J.P., Menis, S., Ota, T., Kalyuzhniy, O., McGuire, A., Sok, D., Huang, P.S., MacPherson, S., Jones, M., Nieusma, T., Mathison, J., Baker, D., Ward, A.B., Burton, D.R., Stamatatos, L., Nemazee, D., Wilson, I.A., Schief, W.R.(2013) Science 340: 711-716
- PubMed: 23539181
- DOI: https://doi.org/10.1126/science.1234150
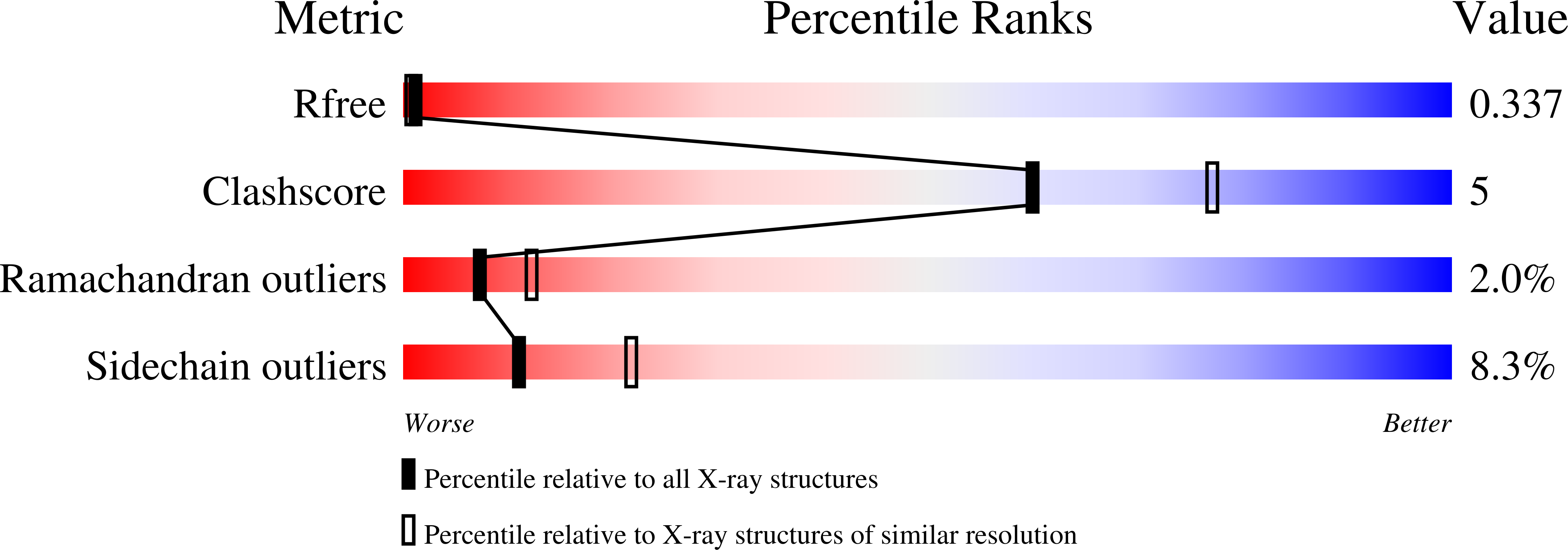
- Primary Citation of Related Structures:
4JPI, 4JPJ, 4JPK - PubMed Abstract:
Vaccine development to induce broadly neutralizing antibodies (bNAbs) against HIV-1 is a global health priority. Potent VRC01-class bNAbs against the CD4 binding site of HIV gp120 have been isolated from HIV-1-infected individuals; however, such bNAbs have not been induced by vaccination. Wild-type gp120 proteins lack detectable affinity for predicted germline precursors of VRC01-class bNAbs, making them poor immunogens to prime a VRC01-class response. We employed computation-guided, in vitro screening to engineer a germline-targeting gp120 outer domain immunogen that binds to multiple VRC01-class bNAbs and germline precursors, and elucidated germline binding crystallographically. When multimerized on nanoparticles, this immunogen (eOD-GT6) activates germline and mature VRC01-class B cells. Thus, eOD-GT6 nanoparticles have promise as a vaccine prime. In principle, germline-targeting strategies could be applied to other epitopes and pathogens.
Organizational Affiliation:
Department of Immunology and Microbial Science, The Scripps Research Institute, La Jolla, CA 92037, USA.